Florentin Bieder
Optimizing Rank for High-Fidelity Implicit Neural Representations
Dec 16, 2025Abstract:Implicit Neural Representations (INRs) based on vanilla Multi-Layer Perceptrons (MLPs) are widely believed to be incapable of representing high-frequency content. This has directed research efforts towards architectural interventions, such as coordinate embeddings or specialized activation functions, to represent high-frequency signals. In this paper, we challenge the notion that the low-frequency bias of vanilla MLPs is an intrinsic, architectural limitation to learn high-frequency content, but instead a symptom of stable rank degradation during training. We empirically demonstrate that regulating the network's rank during training substantially improves the fidelity of the learned signal, rendering even simple MLP architectures expressive. Extensive experiments show that using optimizers like Muon, with high-rank, near-orthogonal updates, consistently enhances INR architectures even beyond simple ReLU MLPs. These substantial improvements hold across a diverse range of domains, including natural and medical images, and novel view synthesis, with up to 9 dB PSNR improvements over the previous state-of-the-art. Our project page, which includes code and experimental results, is available at: (https://muon-inrs.github.io).
VidFuncta: Towards Generalizable Neural Representations for Ultrasound Videos
Jul 29, 2025Abstract:Ultrasound is widely used in clinical care, yet standard deep learning methods often struggle with full video analysis due to non-standardized acquisition and operator bias. We offer a new perspective on ultrasound video analysis through implicit neural representations (INRs). We build on Functa, an INR framework in which each image is represented by a modulation vector that conditions a shared neural network. However, its extension to the temporal domain of medical videos remains unexplored. To address this gap, we propose VidFuncta, a novel framework that leverages Functa to encode variable-length ultrasound videos into compact, time-resolved representations. VidFuncta disentangles each video into a static video-specific vector and a sequence of time-dependent modulation vectors, capturing both temporal dynamics and dataset-level redundancies. Our method outperforms 2D and 3D baselines on video reconstruction and enables downstream tasks to directly operate on the learned 1D modulation vectors. We validate VidFuncta on three public ultrasound video datasets -- cardiac, lung, and breast -- and evaluate its downstream performance on ejection fraction prediction, B-line detection, and breast lesion classification. These results highlight the potential of VidFuncta as a generalizable and efficient representation framework for ultrasound videos. Our code is publicly available under https://github.com/JuliaWolleb/VidFuncta_public.
fastWDM3D: Fast and Accurate 3D Healthy Tissue Inpainting
Jul 17, 2025


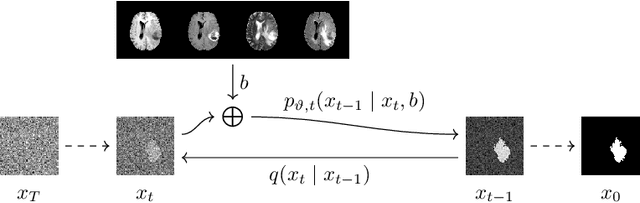
Abstract:Healthy tissue inpainting has significant applications, including the generation of pseudo-healthy baselines for tumor growth models and the facilitation of image registration. In previous editions of the BraTS Local Synthesis of Healthy Brain Tissue via Inpainting Challenge, denoising diffusion probabilistic models (DDPMs) demonstrated qualitatively convincing results but suffered from low sampling speed. To mitigate this limitation, we adapted a 2D image generation approach, combining DDPMs with generative adversarial networks (GANs) and employing a variance-preserving noise schedule, for the task of 3D inpainting. Our experiments showed that the variance-preserving noise schedule and the selected reconstruction losses can be effectively utilized for high-quality 3D inpainting in a few time steps without requiring adversarial training. We applied our findings to a different architecture, a 3D wavelet diffusion model (WDM3D) that does not include a GAN component. The resulting model, denoted as fastWDM3D, obtained a SSIM of 0.8571, a MSE of 0.0079, and a PSNR of 22.26 on the BraTS inpainting test set. Remarkably, it achieved these scores using only two time steps, completing the 3D inpainting process in 1.81 s per image. When compared to other DDPMs used for healthy brain tissue inpainting, our model is up to 800 x faster while still achieving superior performance metrics. Our proposed method, fastWDM3D, represents a promising approach for fast and accurate healthy tissue inpainting. Our code is available at https://github.com/AliciaDurrer/fastWDM3D.
MedFuncta: Modality-Agnostic Representations Based on Efficient Neural Fields
Feb 20, 2025



Abstract:Recent research in medical image analysis with deep learning almost exclusively focuses on grid- or voxel-based data representations. We challenge this common choice by introducing MedFuncta, a modality-agnostic continuous data representation based on neural fields. We demonstrate how to scale neural fields from single instances to large datasets by exploiting redundancy in medical signals and by applying an efficient meta-learning approach with a context reduction scheme. We further address the spectral bias in commonly used SIREN activations, by introducing an $\omega_0$-schedule, improving reconstruction quality and convergence speed. We validate our proposed approach on a large variety of medical signals of different dimensions and modalities (1D: ECG; 2D: Chest X-ray, Retinal OCT, Fundus Camera, Dermatoscope, Colon Histopathology, Cell Microscopy; 3D: Brain MRI, Lung CT) and successfully demonstrate that we can solve relevant downstream tasks on these representations. We additionally release a large-scale dataset of > 550k annotated neural fields to promote research in this direction.
Modeling the Neonatal Brain Development Using Implicit Neural Representations
Aug 16, 2024Abstract:The human brain undergoes rapid development during the third trimester of pregnancy. In this work, we model the neonatal development of the infant brain in this age range. As a basis, we use MR images of preterm- and term-birth neonates from the developing human connectome project (dHCP). We propose a neural network, specifically an implicit neural representation (INR), to predict 2D- and 3D images of varying time points. In order to model a subject-specific development process, it is necessary to disentangle the age from the subjects' identity in the latent space of the INR. We propose two methods, Subject Specific Latent Vectors (SSL) and Stochastic Global Latent Augmentation (SGLA), enabling this disentanglement. We perform an analysis of the results and compare our proposed model to an age-conditioned denoising diffusion model as a baseline. We also show that our method can be applied in a memory-efficient way, which is especially important for 3D data.
Denoising Diffusion Models for 3D Healthy Brain Tissue Inpainting
Mar 21, 2024



Abstract:Monitoring diseases that affect the brain's structural integrity requires automated analysis of magnetic resonance (MR) images, e.g., for the evaluation of volumetric changes. However, many of the evaluation tools are optimized for analyzing healthy tissue. To enable the evaluation of scans containing pathological tissue, it is therefore required to restore healthy tissue in the pathological areas. In this work, we explore and extend denoising diffusion models for consistent inpainting of healthy 3D brain tissue. We modify state-of-the-art 2D, pseudo-3D, and 3D methods working in the image space, as well as 3D latent and 3D wavelet diffusion models, and train them to synthesize healthy brain tissue. Our evaluation shows that the pseudo-3D model performs best regarding the structural-similarity index, peak signal-to-noise ratio, and mean squared error. To emphasize the clinical relevance, we fine-tune this model on data containing synthetic MS lesions and evaluate it on a downstream brain tissue segmentation task, whereby it outperforms the established FMRIB Software Library (FSL) lesion-filling method.
Binary Noise for Binary Tasks: Masked Bernoulli Diffusion for Unsupervised Anomaly Detection
Mar 18, 2024

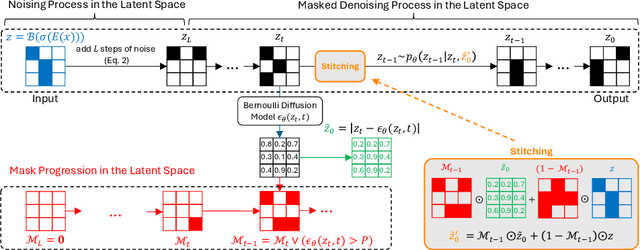

Abstract:The high performance of denoising diffusion models for image generation has paved the way for their application in unsupervised medical anomaly detection. As diffusion-based methods require a lot of GPU memory and have long sampling times, we present a novel and fast unsupervised anomaly detection approach based on latent Bernoulli diffusion models. We first apply an autoencoder to compress the input images into a binary latent representation. Next, a diffusion model that follows a Bernoulli noise schedule is employed to this latent space and trained to restore binary latent representations from perturbed ones. The binary nature of this diffusion model allows us to identify entries in the latent space that have a high probability of flipping their binary code during the denoising process, which indicates out-of-distribution data. We propose a masking algorithm based on these probabilities, which improves the anomaly detection scores. We achieve state-of-the-art performance compared to other diffusion-based unsupervised anomaly detection algorithms while significantly reducing sampling time and memory consumption. The code is available at https://github.com/JuliaWolleb/Anomaly_berdiff.
WDM: 3D Wavelet Diffusion Models for High-Resolution Medical Image Synthesis
Feb 29, 2024Abstract:Due to the three-dimensional nature of CT- or MR-scans, generative modeling of medical images is a particularly challenging task. Existing approaches mostly apply patch-wise, slice-wise, or cascaded generation techniques to fit the high-dimensional data into the limited GPU memory. However, these approaches may introduce artifacts and potentially restrict the model's applicability for certain downstream tasks. This work presents WDM, a wavelet-based medical image synthesis framework that applies a diffusion model on wavelet decomposed images. The presented approach is a simple yet effective way of scaling diffusion models to high resolutions and can be trained on a single 40 GB GPU. Experimental results on BraTS and LIDC-IDRI unconditional image generation at a resolution of $128 \times 128 \times 128$ show state-of-the-art image fidelity (FID) and sample diversity (MS-SSIM) scores compared to GANs, Diffusion Models, and Latent Diffusion Models. Our proposed method is the only one capable of generating high-quality images at a resolution of $256 \times 256 \times 256$.
Diffusion Models for Memory-efficient Processing of 3D Medical Images
Mar 27, 2023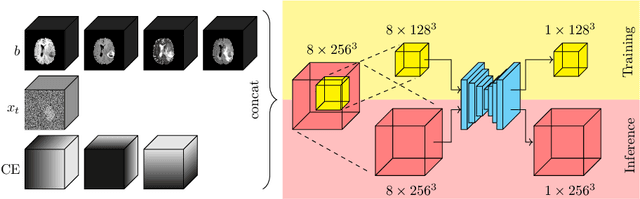

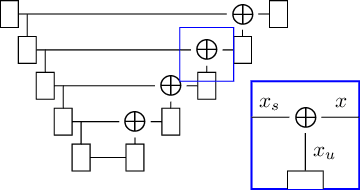
Abstract:Denoising diffusion models have recently achieved state-of-the-art performance in many image-generation tasks. They do, however, require a large amount of computational resources. This limits their application to medical tasks, where we often deal with large 3D volumes, like high-resolution three-dimensional data. In this work, we present a number of different ways to reduce the resource consumption for 3D diffusion models and apply them to a dataset of 3D images. The main contribution of this paper is the memory-efficient patch-based diffusion model \textit{PatchDDM}, which can be applied to the total volume during inference while the training is performed only on patches. While the proposed diffusion model can be applied to any image generation tasks, we evaluate the method on the tumor segmentation task of the BraTS2020 dataset and demonstrate that we can generate meaningful three-dimensional segmentations.
Point Cloud Diffusion Models for Automatic Implant Generation
Mar 14, 2023Abstract:Advances in 3D printing of biocompatible materials make patient-specific implants increasingly popular. The design of these implants is, however, still a tedious and largely manual process. Existing approaches to automate implant generation are mainly based on 3D U-Net architectures on downsampled or patch-wise data, which can result in a loss of detail or contextual information. Following the recent success of Diffusion Probabilistic Models, we propose a novel approach for implant generation based on a combination of 3D point cloud diffusion models and voxelization networks. Due to the stochastic sampling process in our diffusion model, we can propose an ensemble of different implants per defect, from which the physicians can choose the most suitable one. We evaluate our method on the SkullBreak and SkullFix datasets, generating high-quality implants and achieving competitive evaluation scores.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge