Robin Sandkühler
Department of Biomedical Engineering, University of Basel, Basel, Switzerland
GAMER-MRIL identifies Disability-Related Brain Changes in Multiple Sclerosis
Aug 15, 2023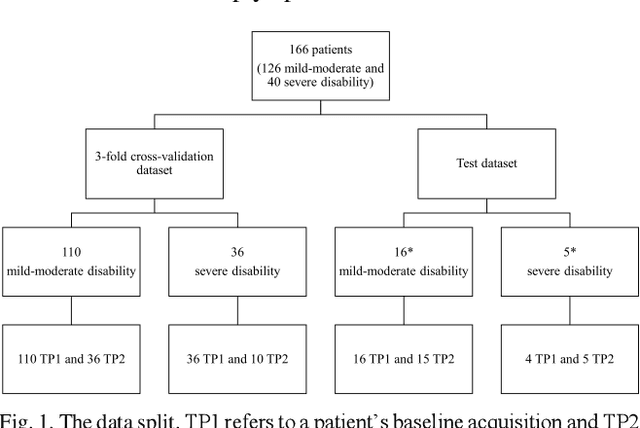
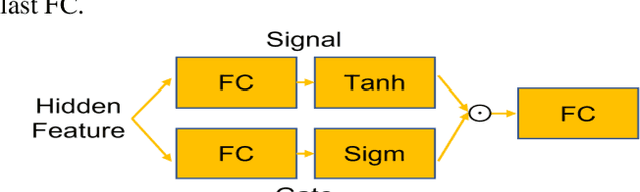
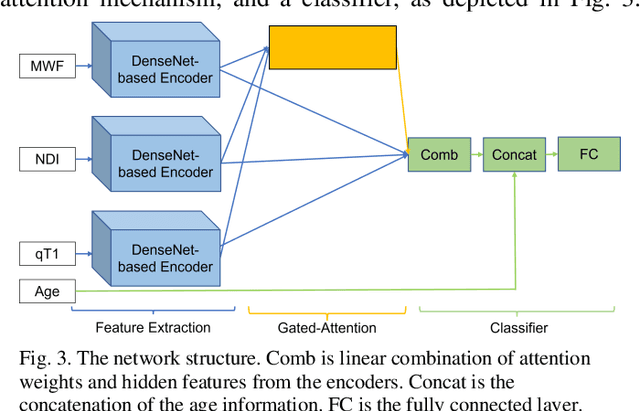
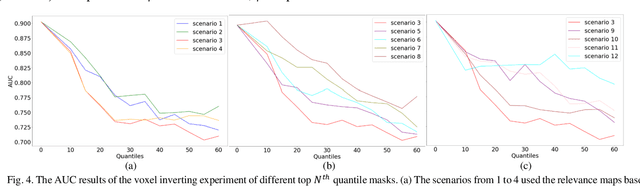
Abstract:Objective: Identifying disability-related brain changes is important for multiple sclerosis (MS) patients. Currently, there is no clear understanding about which pathological features drive disability in single MS patients. In this work, we propose a novel comprehensive approach, GAMER-MRIL, leveraging whole-brain quantitative MRI (qMRI), convolutional neural network (CNN), and an interpretability method from classifying MS patients with severe disability to investigating relevant pathological brain changes. Methods: One-hundred-sixty-six MS patients underwent 3T MRI acquisitions. qMRI informative of microstructural brain properties was reconstructed, including quantitative T1 (qT1), myelin water fraction (MWF), and neurite density index (NDI). To fully utilize the qMRI, GAMER-MRIL extended a gated-attention-based CNN (GAMER-MRI), which was developed to select patch-based qMRI important for a given task/question, to the whole-brain image. To find out disability-related brain regions, GAMER-MRIL modified a structure-aware interpretability method, Layer-wise Relevance Propagation (LRP), to incorporate qMRI. Results: The test performance was AUC=0.885. qT1 was the most sensitive measure related to disability, followed by NDI. The proposed LRP approach obtained more specifically relevant regions than other interpretability methods, including the saliency map, the integrated gradients, and the original LRP. The relevant regions included the corticospinal tract, where average qT1 and NDI significantly correlated with patients' disability scores ($\rho$=-0.37 and 0.44). Conclusion: These results demonstrated that GAMER-MRIL can classify patients with severe disability using qMRI and subsequently identify brain regions potentially important to the integrity of the mobile function. Significance: GAMER-MRIL holds promise for developing biomarkers and increasing clinicians' trust in NN.
Diffusion Models for Memory-efficient Processing of 3D Medical Images
Mar 27, 2023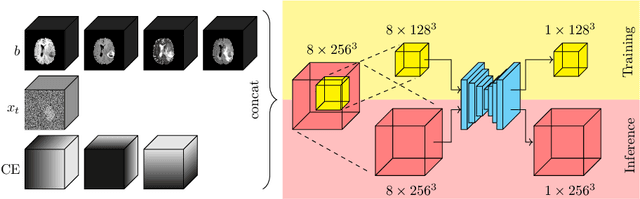

Abstract:Denoising diffusion models have recently achieved state-of-the-art performance in many image-generation tasks. They do, however, require a large amount of computational resources. This limits their application to medical tasks, where we often deal with large 3D volumes, like high-resolution three-dimensional data. In this work, we present a number of different ways to reduce the resource consumption for 3D diffusion models and apply them to a dataset of 3D images. The main contribution of this paper is the memory-efficient patch-based diffusion model \textit{PatchDDM}, which can be applied to the total volume during inference while the training is performed only on patches. While the proposed diffusion model can be applied to any image generation tasks, we evaluate the method on the tumor segmentation task of the BraTS2020 dataset and demonstrate that we can generate meaningful three-dimensional segmentations.
Diffusion Models for Contrast Harmonization of Magnetic Resonance Images
Mar 14, 2023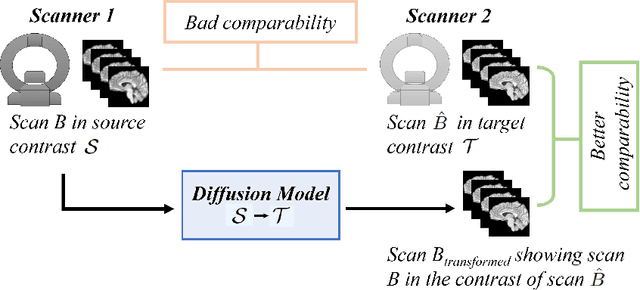
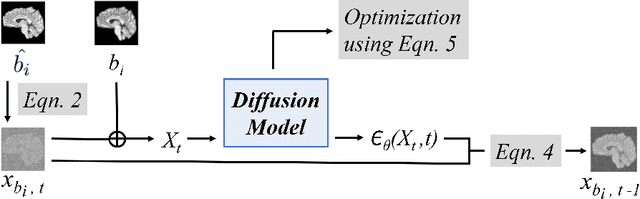
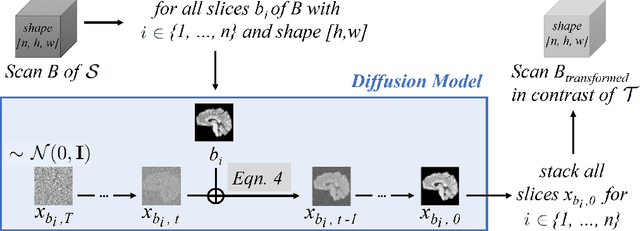

Abstract:Magnetic resonance (MR) images from multiple sources often show differences in image contrast related to acquisition settings or the used scanner type. For long-term studies, longitudinal comparability is essential but can be impaired by these contrast differences, leading to biased results when using automated evaluation tools. This study presents a diffusion model-based approach for contrast harmonization. We use a data set consisting of scans of 18 Multiple Sclerosis patients and 22 healthy controls. Each subject was scanned in two MR scanners of different magnetic field strengths (1.5 T and 3 T), resulting in a paired data set that shows scanner-inherent differences. We map images from the source contrast to the target contrast for both directions, from 3 T to 1.5 T and from 1.5 T to 3 T. As we only want to change the contrast, not the anatomical information, our method uses the original image to guide the image-to-image translation process by adding structural information. The aim is that the mapped scans display increased comparability with scans of the target contrast for downstream tasks. We evaluate this method for the task of segmentation of cerebrospinal fluid, grey matter and white matter. Our method achieves good and consistent results for both directions of the mapping.
Position Regression for Unsupervised Anomaly Detection
Jan 19, 2023



Abstract:In recent years, anomaly detection has become an essential field in medical image analysis. Most current anomaly detection methods for medical images are based on image reconstruction. In this work, we propose a novel anomaly detection approach based on coordinate regression. Our method estimates the position of patches within a volume, and is trained only on data of healthy subjects. During inference, we can detect and localize anomalies by considering the error of the position estimate of a given patch. We apply our method to 3D CT volumes and evaluate it on patients with intracranial haemorrhages and cranial fractures. The results show that our method performs well in detecting these anomalies. Furthermore, we show that our method requires less memory than comparable approaches that involve image reconstruction. This is highly relevant for processing large 3D volumes, for instance, CT or MRI scans.
The Swiss Army Knife for Image-to-Image Translation: Multi-Task Diffusion Models
Apr 06, 2022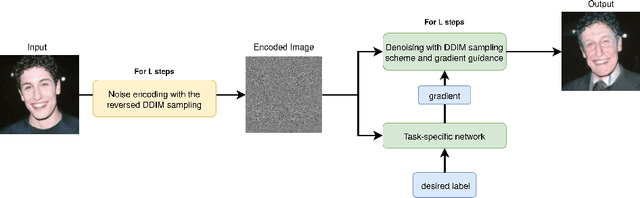



Abstract:Recently, diffusion models were applied to a wide range of image analysis tasks. We build on a method for image-to-image translation using denoising diffusion implicit models and include a regression problem and a segmentation problem for guiding the image generation to the desired output. The main advantage of our approach is that the guidance during the denoising process is done by an external gradient. Consequently, the diffusion model does not need to be retrained for the different tasks on the same dataset. We apply our method to simulate the aging process on facial photos using a regression task, as well as on a brain magnetic resonance (MR) imaging dataset for the simulation of brain tumor growth. Furthermore, we use a segmentation model to inpaint tumors at the desired location in healthy slices of brain MR images. We achieve convincing results for all problems.
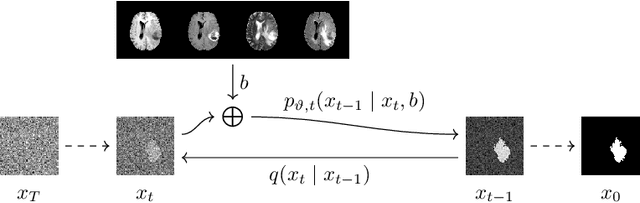
Diffusion Models for Medical Anomaly Detection
Mar 08, 2022



Abstract:In medical applications, weakly supervised anomaly detection methods are of great interest, as only image-level annotations are required for training. Current anomaly detection methods mainly rely on generative adversarial networks or autoencoder models. Those models are often complicated to train or have difficulties to preserve fine details in the image. We present a novel weakly supervised anomaly detection method based on denoising diffusion implicit models. We combine the deterministic iterative noising and denoising scheme with classifier guidance for image-to-image translation between diseased and healthy subjects. Our method generates very detailed anomaly maps without the need for a complex training procedure. We evaluate our method on the BRATS2020 dataset for brain tumor detection and the CheXpert dataset for detecting pleural effusions.
Diffusion Models for Implicit Image Segmentation Ensembles
Dec 27, 2021



Abstract:Diffusion models have shown impressive performance for generative modelling of images. In this paper, we present a novel semantic segmentation method based on diffusion models. By modifying the training and sampling scheme, we show that diffusion models can perform lesion segmentation of medical images. To generate an image specific segmentation, we train the model on the ground truth segmentation, and use the image as a prior during training and in every step during the sampling process. With the given stochastic sampling process, we can generate a distribution of segmentation masks. This property allows us to compute pixel-wise uncertainty maps of the segmentation, and allows an implicit ensemble of segmentations that increases the segmentation performance. We evaluate our method on the BRATS2020 dataset for brain tumor segmentation. Compared to state-of-the-art segmentation models, our approach yields good segmentation results and, additionally, detailed uncertainty maps.
Learn to Ignore: Domain Adaptation for Multi-Site MRI Analysis
Oct 13, 2021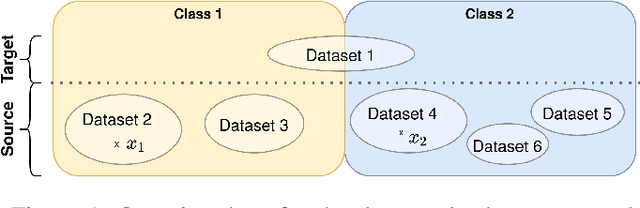

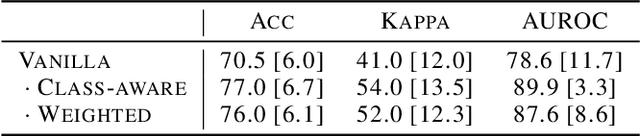
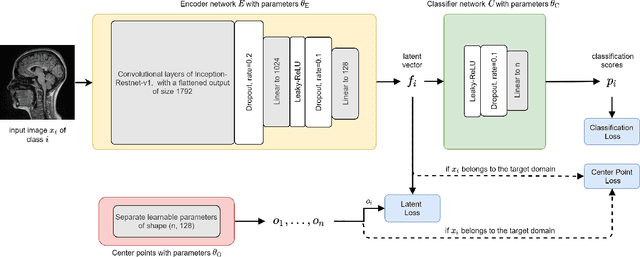
Abstract:Limited availability of large image datasets is a major issue in the development of accurate and generalizable machine learning methods in medicine. The limitations in the amount of data are mainly due to the use of different acquisition protocols, different hardware, and data privacy. At the same time, training a classification model on a small dataset leads to a poor generalization quality of the model. To overcome this issue, a combination of various image datasets of different provenance is often used, e.g., multi-site studies. However, if an additional dataset does not include all classes of the task, the learning of the classification model can be biased to the device or place of acquisition. This is especially the case for Magnetic Resonance (MR) images, where different MR scanners introduce a bias that limits the performance of the model. In this paper, we present a novel method that learns to ignore the scanner-related features present in the images, while learning features relevant for the classification task. We focus on a real-world scenario, where only a small dataset provides images of all classes. We exploit this circumstance by introducing specific additional constraints on the latent space, which lead the focus on disease-related rather than scanner-specific features. Our method Learn to Ignore outperforms state-of-the-art domain adaptation methods on a multi-site MRI dataset on a classification task between Multiple Sclerosis patients and healthy subjects.
MRI lung lobe segmentation in pediatric cystic fibrosis patients using a recurrent neural network trained with publicly accessible CT datasets
Aug 31, 2021



Abstract:Purpose: To introduce a widely applicable workflow for pulmonary lobe segmentation of MR images using a recurrent neural network (RNN) trained with chest computed tomography (CT) datasets. The feasibility is demonstrated for 2D coronal ultra-fast balanced steady-state free precession (ufSSFP) MRI. Methods: Lung lobes of 250 publicly accessible CT datasets of adults were segmented with an open-source CT-specific algorithm. To match 2D ufSSFP MRI data of pediatric patients, both CT data and segmentations were translated into pseudo-MR images, masked to suppress anatomy outside the lung. Network-1 was trained with pseudo-MR images and lobe segmentations, and applied to 1000 masked ufSSFP images to predict lobe segmentations. These outputs were directly used as targets to train Network-2 and Network-3 with non-masked ufSSFP data as inputs, and an additional whole-lung mask as input for Network-2. Network predictions were compared to reference manual lobe segmentations of ufSSFP data in twenty pediatric cystic fibrosis patients. Manual lobe segmentations were performed by splitting available whole-lung segmentations into lobes. Results: Network-1 was able to segment the lobes of ufSSFP images, and Network-2 and Network-3 further increased segmentation accuracy and robustness. The average all-lobe Dice similarity coefficients were 95.0$\pm$2.3 (mean$\pm$pooled SD [%]), 96.4$\pm$1.2, 93.0$\pm$1.8, and the average median Hausdorff distances were 6.1$\pm$0.9 (mean$\pm$SD [mm]), 5.3$\pm$1.1, 7.1$\pm$1.3, for Network-1, Network-2, and Network-3, respectively. Conclusions: RNN lung lobe segmentation of 2D ufSSFP imaging is feasible, in good agreement with manual segmentations. The proposed workflow might provide rapid access to automated lobe segmentations for various lung MRI examinations and quantitative analyses.
Comparison of Methods Generalizing Max- and Average-Pooling
Mar 02, 2021



Abstract:Max- and average-pooling are the most popular pooling methods for downsampling in convolutional neural networks. In this paper, we compare different pooling methods that generalize both max- and average-pooling. Furthermore, we propose another method based on a smooth approximation of the maximum function and put it into context with related methods. For the comparison, we use a VGG16 image classification network and train it on a large dataset of natural high-resolution images (Google Open Images v5). The results show that none of the more sophisticated methods perform significantly better in this classification task than standard max- or average-pooling.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge