Özgür Yaldizli
Denoising Diffusion Models for 3D Healthy Brain Tissue Inpainting
Mar 21, 2024



Abstract:Monitoring diseases that affect the brain's structural integrity requires automated analysis of magnetic resonance (MR) images, e.g., for the evaluation of volumetric changes. However, many of the evaluation tools are optimized for analyzing healthy tissue. To enable the evaluation of scans containing pathological tissue, it is therefore required to restore healthy tissue in the pathological areas. In this work, we explore and extend denoising diffusion models for consistent inpainting of healthy 3D brain tissue. We modify state-of-the-art 2D, pseudo-3D, and 3D methods working in the image space, as well as 3D latent and 3D wavelet diffusion models, and train them to synthesize healthy brain tissue. Our evaluation shows that the pseudo-3D model performs best regarding the structural-similarity index, peak signal-to-noise ratio, and mean squared error. To emphasize the clinical relevance, we fine-tune this model on data containing synthetic MS lesions and evaluate it on a downstream brain tissue segmentation task, whereby it outperforms the established FMRIB Software Library (FSL) lesion-filling method.
Diffusion Models for Contrast Harmonization of Magnetic Resonance Images
Mar 14, 2023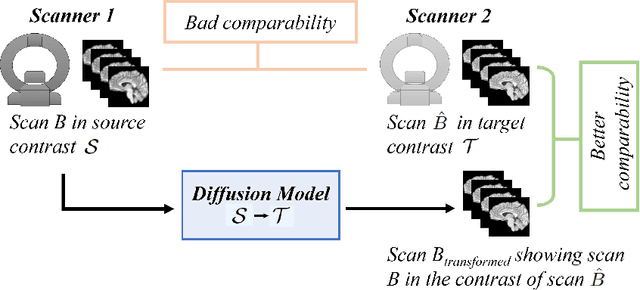
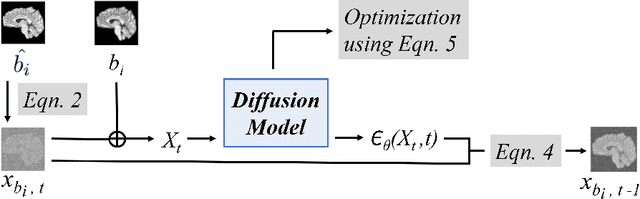
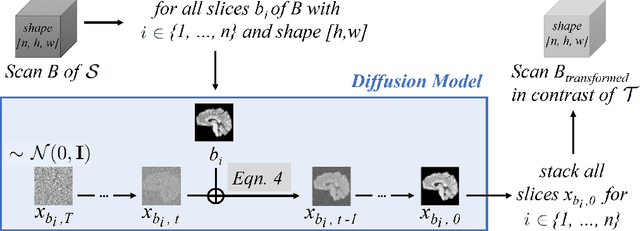

Abstract:Magnetic resonance (MR) images from multiple sources often show differences in image contrast related to acquisition settings or the used scanner type. For long-term studies, longitudinal comparability is essential but can be impaired by these contrast differences, leading to biased results when using automated evaluation tools. This study presents a diffusion model-based approach for contrast harmonization. We use a data set consisting of scans of 18 Multiple Sclerosis patients and 22 healthy controls. Each subject was scanned in two MR scanners of different magnetic field strengths (1.5 T and 3 T), resulting in a paired data set that shows scanner-inherent differences. We map images from the source contrast to the target contrast for both directions, from 3 T to 1.5 T and from 1.5 T to 3 T. As we only want to change the contrast, not the anatomical information, our method uses the original image to guide the image-to-image translation process by adding structural information. The aim is that the mapped scans display increased comparability with scans of the target contrast for downstream tasks. We evaluate this method for the task of segmentation of cerebrospinal fluid, grey matter and white matter. Our method achieves good and consistent results for both directions of the mapping.
Learn to Ignore: Domain Adaptation for Multi-Site MRI Analysis
Oct 13, 2021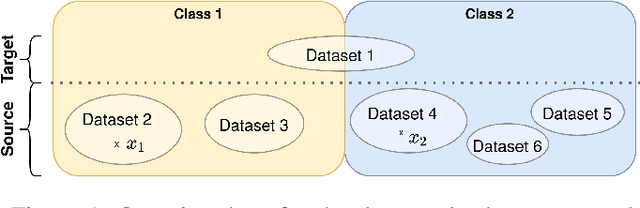

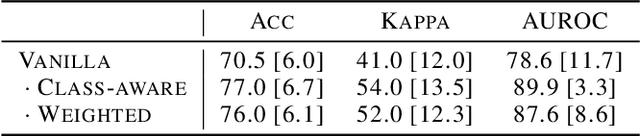
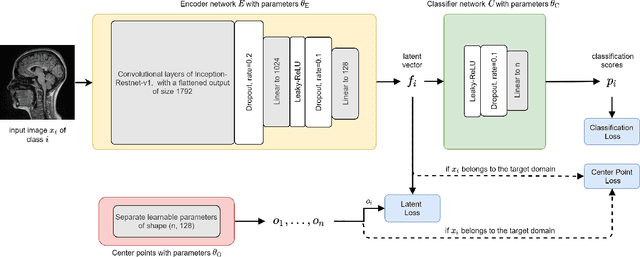
Abstract:Limited availability of large image datasets is a major issue in the development of accurate and generalizable machine learning methods in medicine. The limitations in the amount of data are mainly due to the use of different acquisition protocols, different hardware, and data privacy. At the same time, training a classification model on a small dataset leads to a poor generalization quality of the model. To overcome this issue, a combination of various image datasets of different provenance is often used, e.g., multi-site studies. However, if an additional dataset does not include all classes of the task, the learning of the classification model can be biased to the device or place of acquisition. This is especially the case for Magnetic Resonance (MR) images, where different MR scanners introduce a bias that limits the performance of the model. In this paper, we present a novel method that learns to ignore the scanner-related features present in the images, while learning features relevant for the classification task. We focus on a real-world scenario, where only a small dataset provides images of all classes. We exploit this circumstance by introducing specific additional constraints on the latent space, which lead the focus on disease-related rather than scanner-specific features. Our method Learn to Ignore outperforms state-of-the-art domain adaptation methods on a multi-site MRI dataset on a classification task between Multiple Sclerosis patients and healthy subjects.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge