Henning Muller
Multi-Class Segmentation of Aortic Branches and Zones in Computed Tomography Angiography: The AortaSeg24 Challenge
Feb 07, 2025



Abstract:Multi-class segmentation of the aorta in computed tomography angiography (CTA) scans is essential for diagnosing and planning complex endovascular treatments for patients with aortic dissections. However, existing methods reduce aortic segmentation to a binary problem, limiting their ability to measure diameters across different branches and zones. Furthermore, no open-source dataset is currently available to support the development of multi-class aortic segmentation methods. To address this gap, we organized the AortaSeg24 MICCAI Challenge, introducing the first dataset of 100 CTA volumes annotated for 23 clinically relevant aortic branches and zones. This dataset was designed to facilitate both model development and validation. The challenge attracted 121 teams worldwide, with participants leveraging state-of-the-art frameworks such as nnU-Net and exploring novel techniques, including cascaded models, data augmentation strategies, and custom loss functions. We evaluated the submitted algorithms using the Dice Similarity Coefficient (DSC) and Normalized Surface Distance (NSD), highlighting the approaches adopted by the top five performing teams. This paper presents the challenge design, dataset details, evaluation metrics, and an in-depth analysis of the top-performing algorithms. The annotated dataset, evaluation code, and implementations of the leading methods are publicly available to support further research. All resources can be accessed at https://aortaseg24.grand-challenge.org.
Structural-Based Uncertainty in Deep Learning Across Anatomical Scales: Analysis in White Matter Lesion Segmentation
Nov 15, 2023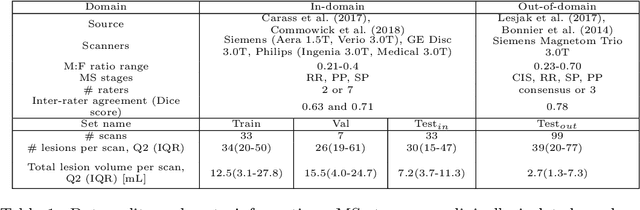


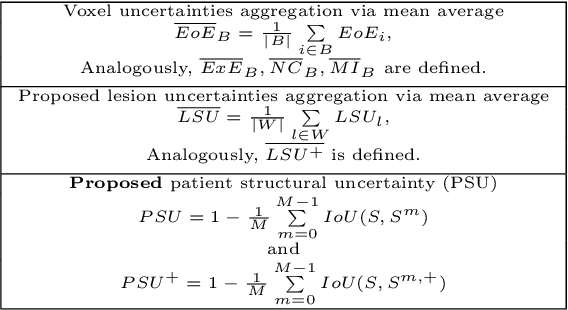
Abstract:This paper explores uncertainty quantification (UQ) as an indicator of the trustworthiness of automated deep-learning (DL) tools in the context of white matter lesion (WML) segmentation from magnetic resonance imaging (MRI) scans of multiple sclerosis (MS) patients. Our study focuses on two principal aspects of uncertainty in structured output segmentation tasks. Firstly, we postulate that a good uncertainty measure should indicate predictions likely to be incorrect with high uncertainty values. Second, we investigate the merit of quantifying uncertainty at different anatomical scales (voxel, lesion, or patient). We hypothesize that uncertainty at each scale is related to specific types of errors. Our study aims to confirm this relationship by conducting separate analyses for in-domain and out-of-domain settings. Our primary methodological contributions are (i) the development of novel measures for quantifying uncertainty at lesion and patient scales, derived from structural prediction discrepancies, and (ii) the extension of an error retention curve analysis framework to facilitate the evaluation of UQ performance at both lesion and patient scales. The results from a multi-centric MRI dataset of 172 patients demonstrate that our proposed measures more effectively capture model errors at the lesion and patient scales compared to measures that average voxel-scale uncertainty values. We provide the UQ protocols code at https://github.com/Medical-Image-Analysis-Laboratory/MS_WML_uncs.
Disentangling Neuron Representations with Concept Vectors
Apr 19, 2023



Abstract:Mechanistic interpretability aims to understand how models store representations by breaking down neural networks into interpretable units. However, the occurrence of polysemantic neurons, or neurons that respond to multiple unrelated features, makes interpreting individual neurons challenging. This has led to the search for meaningful vectors, known as concept vectors, in activation space instead of individual neurons. The main contribution of this paper is a method to disentangle polysemantic neurons into concept vectors encapsulating distinct features. Our method can search for fine-grained concepts according to the user's desired level of concept separation. The analysis shows that polysemantic neurons can be disentangled into directions consisting of linear combinations of neurons. Our evaluations show that the concept vectors found encode coherent, human-understandable features.
Tackling Bias in the Dice Similarity Coefficient: Introducing nDSC for White Matter Lesion Segmentation
Feb 10, 2023Abstract:The development of automatic segmentation techniques for medical imaging tasks requires assessment metrics to fairly judge and rank such approaches on benchmarks. The Dice Similarity Coefficient (DSC) is a popular choice for comparing the agreement between the predicted segmentation against a ground-truth mask. However, the DSC metric has been shown to be biased to the occurrence rate of the positive class in the ground-truth, and hence should be considered in combination with other metrics. This work describes a detailed analysis of the recently proposed normalised Dice Similarity Coefficient (nDSC) for binary segmentation tasks as an adaptation of DSC which scales the precision at a fixed recall rate to tackle this bias. White matter lesion segmentation on magnetic resonance images of multiple sclerosis patients is selected as a case study task to empirically assess the suitability of nDSC. We validate the normalised DSC using two different models across 59 subject scans with a wide range of lesion loads. It is found that the nDSC is less biased than DSC with lesion load on standard white matter lesion segmentation benchmarks measured using standard rank correlation coefficients. An implementation of nDSC is made available at: https://github.com/NataliiaMolch/nDSC .
Unsupervised Method for Intra-patient Registration of Brain Magnetic Resonance Images based on Objective Function Weighting by Inverse Consistency: Contribution to the BraTS-Reg Challenge
Nov 14, 2022Abstract:Registration of brain scans with pathologies is difficult, yet important research area. The importance of this task motivated researchers to organize the BraTS-Reg challenge, jointly with IEEE ISBI 2022 and MICCAI 2022 conferences. The organizers introduced the task of aligning pre-operative to follow-up magnetic resonance images of glioma. The main difficulties are connected with the missing data leading to large, nonrigid, and noninvertible deformations. In this work, we describe our contributions to both the editions of the BraTS-Reg challenge. The proposed method is based on combined deep learning and instance optimization approaches. First, the instance optimization enriches the state-of-the-art LapIRN method to improve the generalizability and fine-details preservation. Second, an additional objective function weighting is introduced, based on the inverse consistency. The proposed method is fully unsupervised and exhibits high registration quality and robustness. The quantitative results on the external validation set are: (i) IEEE ISBI 2022 edition: 1.85, and 0.86, (ii) MICCAI 2022 edition: 1.71, and 0.86, in terms of the mean of median absolute error and robustness respectively. The method scored the 1st place during the IEEE ISBI 2022 version of the challenge and the 3rd place during the MICCAI 2022. Future work could transfer the inverse consistency-based weighting directly into the deep network training.
Novel structural-scale uncertainty measures and error retention curves: application to multiple sclerosis
Nov 11, 2022Abstract:This paper focuses on the uncertainty estimation for white matter lesions (WML) segmentation in magnetic resonance imaging (MRI). On one side, voxel-scale segmentation errors cause the erroneous delineation of the lesions; on the other side, lesion-scale detection errors lead to wrong lesion counts. Both of these factors are clinically relevant for the assessment of multiple sclerosis patients. This work aims to compare the ability of different voxel- and lesion-scale uncertainty measures to capture errors related to segmentation and lesion detection, respectively. Our main contributions are (i) proposing new measures of lesion-scale uncertainty that do not utilise voxel-scale uncertainties; (ii) extending an error retention curves analysis framework for evaluation of lesion-scale uncertainty measures. Our results obtained on the multi-center testing set of 58 patients demonstrate that the proposed lesion-scale measure achieves the best performance among the analysed measures. All code implementations are provided at https://github.com/NataliiaMolch/MS_WML_uncs
Guiding CNNs towards Relevant Concepts by Multi-task and Adversarial Learning
Aug 04, 2020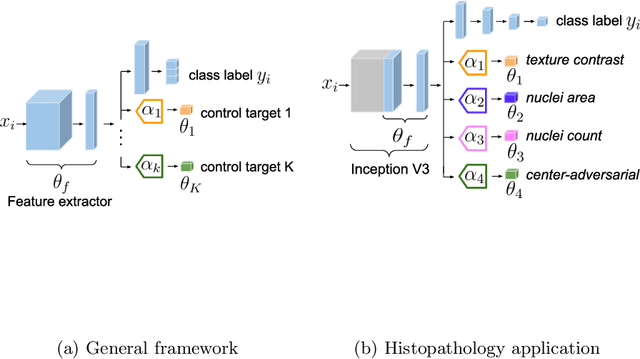
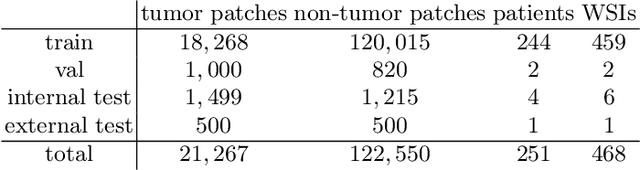
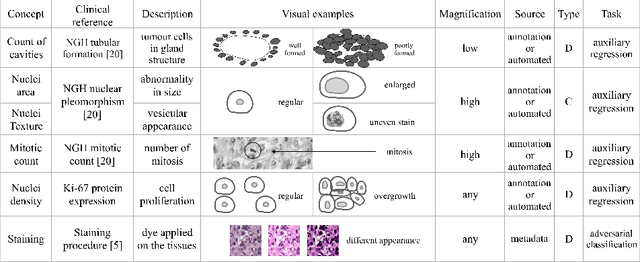
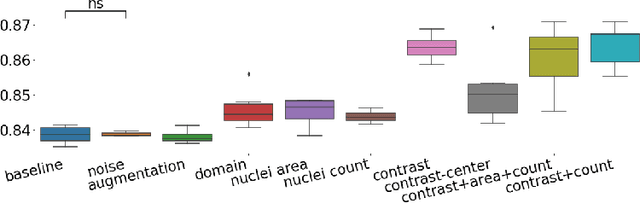
Abstract:The opaqueness of deep learning limits its deployment in critical application scenarios such as cancer grading in medical images. In this paper, a framework for guiding CNN training is built on top of successful existing techniques of hard parameter sharing, with the main goal of explicitly introducing expert knowledge in the training objectives. The learning process is guided by identifying concepts that are relevant or misleading for the task. Relevant concepts are encouraged to appear in the representation through multi-task learning. Undesired and misleading concepts are discouraged by a gradient reversal operation. In this way, a shift in the deep representations can be corrected to match the clinicians' assumptions. The application on breast lymph nodes histopathology data from the Camelyon challenge shows a significant increase in the generalization performance on unseen patients (from 0.839 to 0.864 average AUC, $\text{p-value} = 0,0002$) when the internal representations are controlled by prior knowledge regarding the acquisition center and visual features of the tissue. The code will be shared for reproducibility on our GitHub repository.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge