Mathieu Hatt
LaTIM
MedShapeNet -- A Large-Scale Dataset of 3D Medical Shapes for Computer Vision
Sep 12, 2023



Abstract:We present MedShapeNet, a large collection of anatomical shapes (e.g., bones, organs, vessels) and 3D surgical instrument models. Prior to the deep learning era, the broad application of statistical shape models (SSMs) in medical image analysis is evidence that shapes have been commonly used to describe medical data. Nowadays, however, state-of-the-art (SOTA) deep learning algorithms in medical imaging are predominantly voxel-based. In computer vision, on the contrary, shapes (including, voxel occupancy grids, meshes, point clouds and implicit surface models) are preferred data representations in 3D, as seen from the numerous shape-related publications in premier vision conferences, such as the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), as well as the increasing popularity of ShapeNet (about 51,300 models) and Princeton ModelNet (127,915 models) in computer vision research. MedShapeNet is created as an alternative to these commonly used shape benchmarks to facilitate the translation of data-driven vision algorithms to medical applications, and it extends the opportunities to adapt SOTA vision algorithms to solve critical medical problems. Besides, the majority of the medical shapes in MedShapeNet are modeled directly on the imaging data of real patients, and therefore it complements well existing shape benchmarks comprising of computer-aided design (CAD) models. MedShapeNet currently includes more than 100,000 medical shapes, and provides annotations in the form of paired data. It is therefore also a freely available repository of 3D models for extended reality (virtual reality - VR, augmented reality - AR, mixed reality - MR) and medical 3D printing. This white paper describes in detail the motivations behind MedShapeNet, the shape acquisition procedures, the use cases, as well as the usage of the online shape search portal: https://medshapenet.ikim.nrw/
Joint nnU-Net and Radiomics Approaches for Segmentation and Prognosis of Head and Neck Cancers with PET/CT images
Nov 18, 2022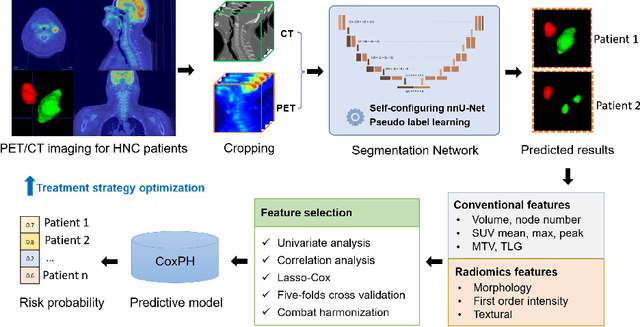
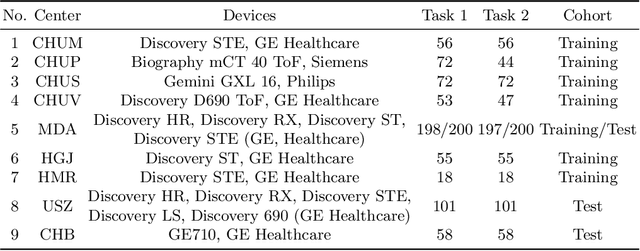
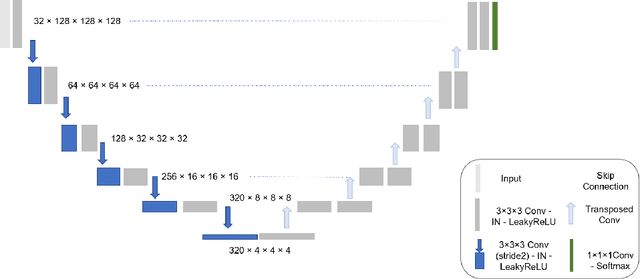
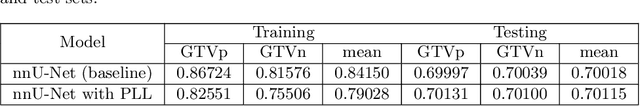
Abstract:Automatic segmentation of head and neck cancer (HNC) tumors and lymph nodes plays a crucial role in the optimization treatment strategy and prognosis analysis. This study aims to employ nnU-Net for automatic segmentation and radiomics for recurrence-free survival (RFS) prediction using pretreatment PET/CT images in multi-center HNC cohort. A multi-center HNC dataset with 883 patients (524 patients for training, 359 for testing) was provided in HECKTOR 2022. A bounding box of the extended oropharyngeal region was retrieved for each patient with fixed size of 224 x 224 x 224 $mm^{3}$. Then 3D nnU-Net architecture was adopted to automatic segmentation of primary tumor and lymph nodes synchronously.Based on predicted segmentation, ten conventional features and 346 standardized radiomics features were extracted for each patient. Three prognostic models were constructed containing conventional and radiomics features alone, and their combinations by multivariate CoxPH modelling. The statistical harmonization method, ComBat, was explored towards reducing multicenter variations. Dice score and C-index were used as evaluation metrics for segmentation and prognosis task, respectively. For segmentation task, we achieved mean dice score around 0.701 for primary tumor and lymph nodes by 3D nnU-Net. For prognostic task, conventional and radiomics models obtained the C-index of 0.658 and 0.645 in the test set, respectively, while the combined model did not improve the prognostic performance with the C-index of 0.648.
Evaluation of importance estimators in deep learning classifiers for Computed Tomography
Sep 30, 2022Abstract:Deep learning has shown superb performance in detecting objects and classifying images, ensuring a great promise for analyzing medical imaging. Translating the success of deep learning to medical imaging, in which doctors need to understand the underlying process, requires the capability to interpret and explain the prediction of neural networks. Interpretability of deep neural networks often relies on estimating the importance of input features (e.g., pixels) with respect to the outcome (e.g., class probability). However, a number of importance estimators (also known as saliency maps) have been developed and it is unclear which ones are more relevant for medical imaging applications. In the present work, we investigated the performance of several importance estimators in explaining the classification of computed tomography (CT) images by a convolutional deep network, using three distinct evaluation metrics. First, the model-centric fidelity measures a decrease in the model accuracy when certain inputs are perturbed. Second, concordance between importance scores and the expert-defined segmentation masks is measured on a pixel level by a receiver operating characteristic (ROC) curves. Third, we measure a region-wise overlap between a XRAI-based map and the segmentation mask by Dice Similarity Coefficients (DSC). Overall, two versions of SmoothGrad topped the fidelity and ROC rankings, whereas both Integrated Gradients and SmoothGrad excelled in DSC evaluation. Interestingly, there was a critical discrepancy between model-centric (fidelity) and human-centric (ROC and DSC) evaluation. Expert expectation and intuition embedded in segmentation maps does not necessarily align with how the model arrived at its prediction. Understanding this difference in interpretability would help harnessing the power of deep learning in medicine.
* 4th International Workshop on EXplainable and TRAnsparent AI and Multi-Agent Systems (EXTRAAMAS 2022) - International Conference on Autonomous Agents and Multi-Agent Systems (AAMAS)
Overview of the HECKTOR Challenge at MICCAI 2021: Automatic Head and Neck Tumor Segmentation and Outcome Prediction in PET/CT Images
Jan 11, 2022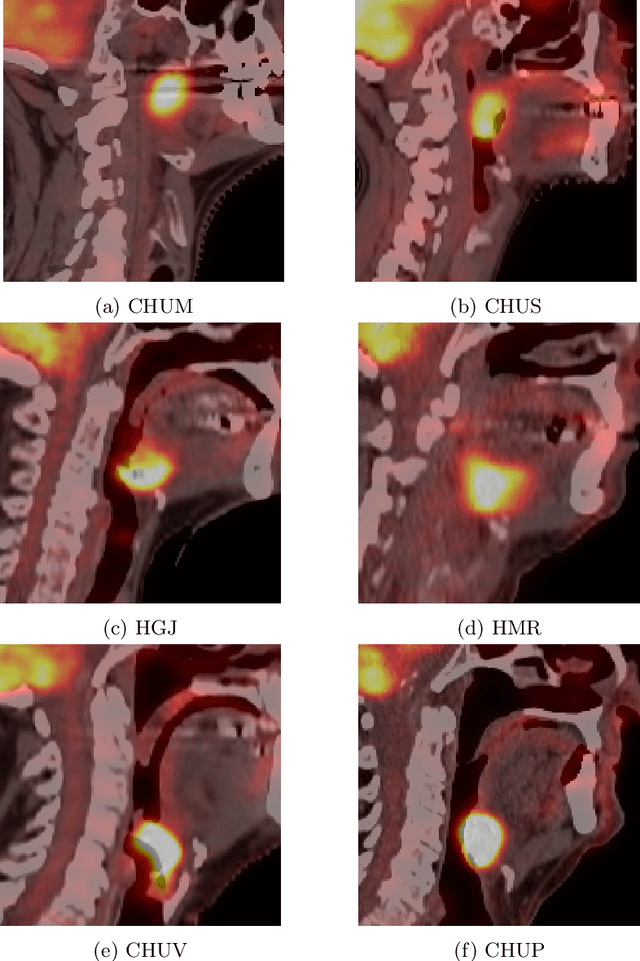
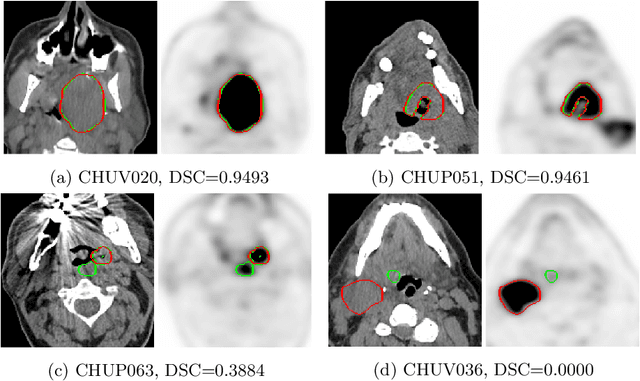
Abstract:This paper presents an overview of the second edition of the HEad and neCK TumOR (HECKTOR) challenge, organized as a satellite event of the 24th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) 2021. The challenge is composed of three tasks related to the automatic analysis of PET/CT images for patients with Head and Neck cancer (H&N), focusing on the oropharynx region. Task 1 is the automatic segmentation of H&N primary Gross Tumor Volume (GTVt) in FDG-PET/CT images. Task 2 is the automatic prediction of Progression Free Survival (PFS) from the same FDG-PET/CT. Finally, Task 3 is the same as Task 2 with ground truth GTVt annotations provided to the participants. The data were collected from six centers for a total of 325 images, split into 224 training and 101 testing cases. The interest in the challenge was highlighted by the important participation with 103 registered teams and 448 result submissions. The best methods obtained a Dice Similarity Coefficient (DSC) of 0.7591 in the first task, and a Concordance index (C-index) of 0.7196 and 0.6978 in Tasks 2 and 3, respectively. In all tasks, simplicity of the approach was found to be key to ensure generalization performance. The comparison of the PFS prediction performance in Tasks 2 and 3 suggests that providing the GTVt contour was not crucial to achieve best results, which indicates that fully automatic methods can be used. This potentially obviates the need for GTVt contouring, opening avenues for reproducible and large scale radiomics studies including thousands potential subjects.
Squeeze-and-Excitation Normalization for Automated Delineation of Head and Neck Primary Tumors in Combined PET and CT Images
Feb 20, 2021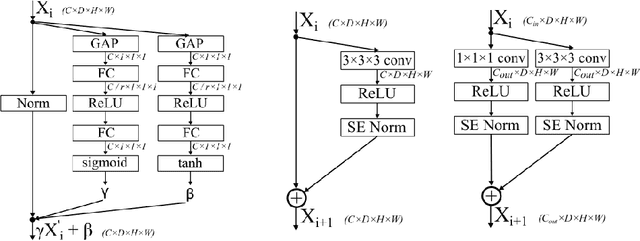
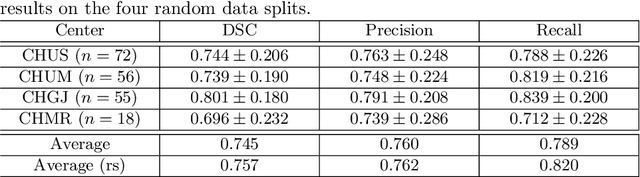
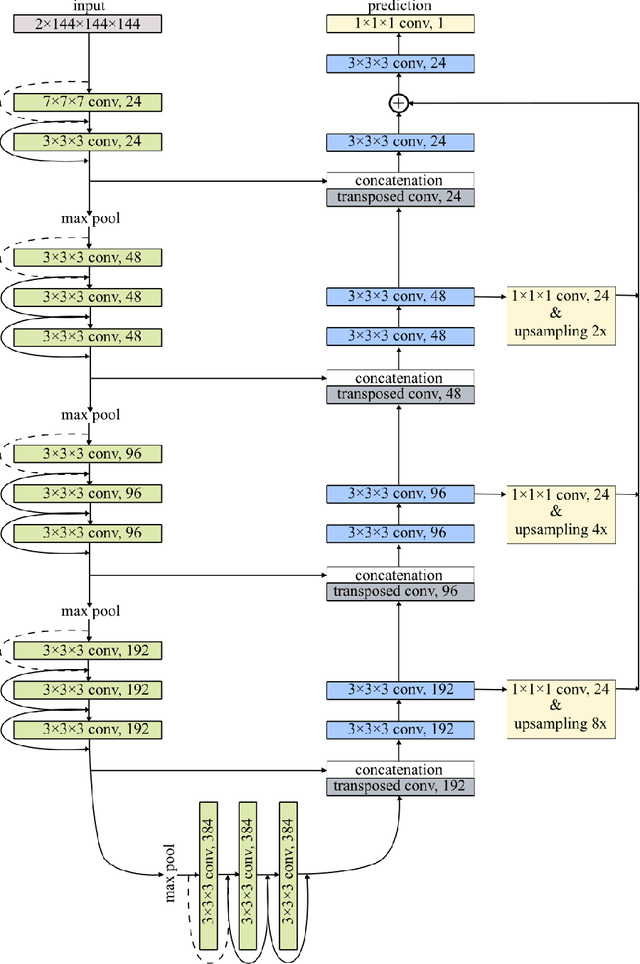

Abstract:Development of robust and accurate fully automated methods for medical image segmentation is crucial in clinical practice and radiomics studies. In this work, we contributed an automated approach for Head and Neck (H&N) primary tumor segmentation in combined positron emission tomography / computed tomography (PET/CT) images in the context of the MICCAI 2020 Head and Neck Tumor segmentation challenge (HECKTOR). Our model was designed on the U-Net architecture with residual layers and supplemented with Squeeze-and-Excitation Normalization. The described method achieved competitive results in cross-validation (DSC 0.745, precision 0.760, recall 0.789) performed on different centers, as well as on the test set (DSC 0.759, precision 0.833, recall 0.740) that allowed us to win first prize in the HECKTOR challenge among 21 participating teams. The full implementation based on PyTorch and the trained models are available at https://github.com/iantsen/hecktor
* 7 pages, 2 figures, 2 tables
Next Generation Radiogenomics Sequencing for Prediction of EGFR and KRAS Mutation Status in NSCLC Patients Using Multimodal Imaging and Machine Learning Approaches
Jul 03, 2019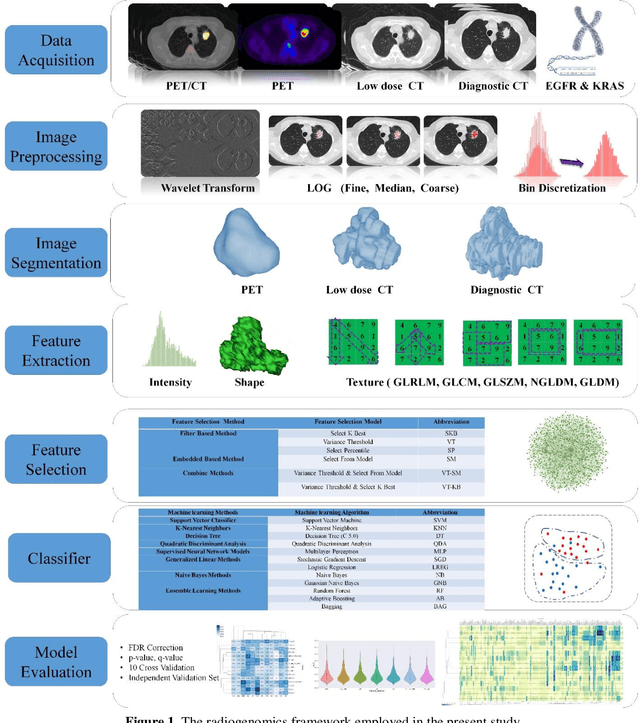
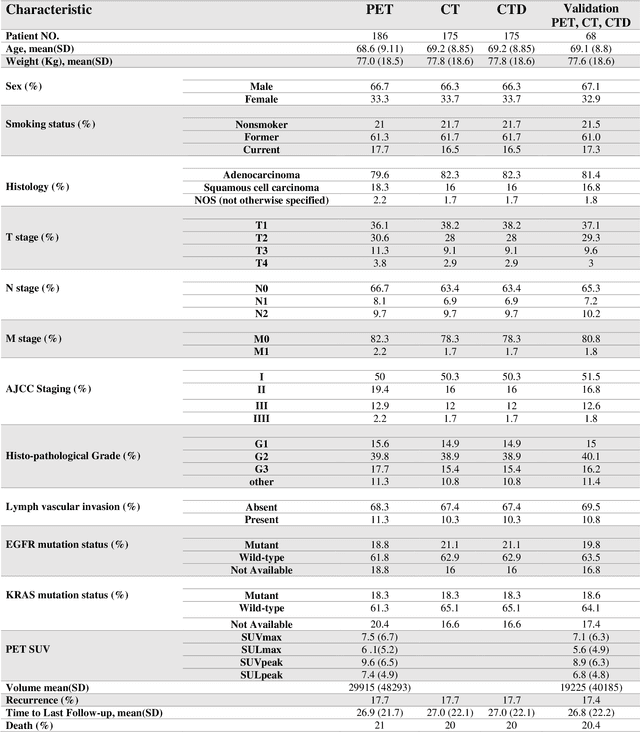

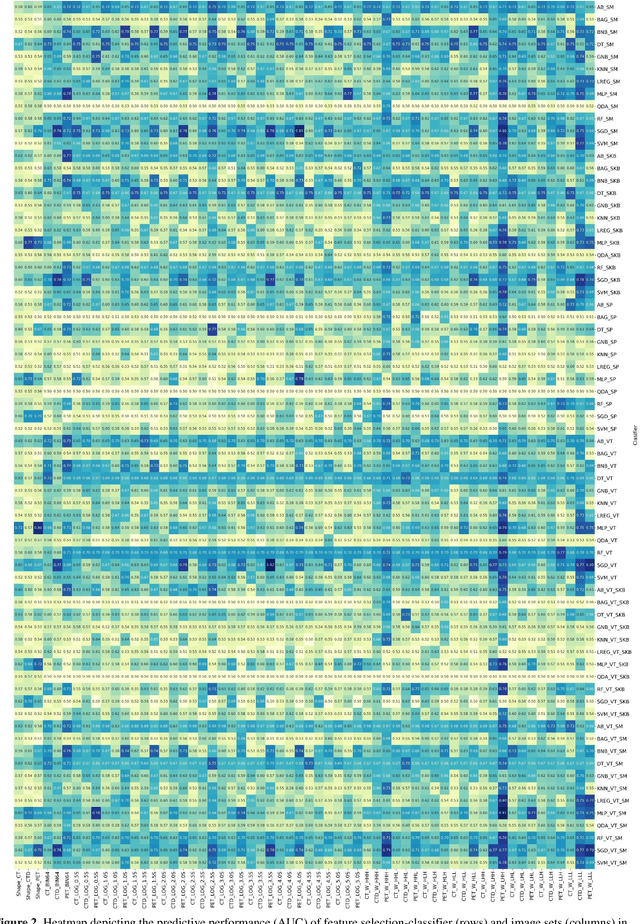
Abstract:Aim: In the present work, we aimed to evaluate a comprehensive radiomics framework that enabled prediction of EGFR and KRAS mutation status in NSCLC cancer patients based on PET and CT multi-modalities radiomic features and machine learning (ML) algorithms. Methods: Our study involved 211 NSCLC cancer patient with PET and CTD images. More than twenty thousand radiomic features from different image-feature sets were extracted Feature value was normalized to obtain Z-scores, followed by student t-test students for comparison, high correlated features were eliminated and the False discovery rate (FDR) correction were performed Six feature selection methods and twelve classifiers were used to predict gene status in patient and model evaluation was reported on independent validation sets (68 patients). Results: The best predictive power of conventional PET parameters was achieved by SUVpeak (AUC: 0.69, P-value = 0.0002) and MTV (AUC: 0.55, P-value = 0.0011) for EGFR and KRAS, respectively. Univariate analysis of radiomics features improved prediction power up to AUC: 75 (q-value: 0.003, Short Run Emphasis feature of GLRLM from LOG preprocessed image of PET with sigma value 1.5) and AUC: 0.71 (q-value 0.00005, The Large Dependence Low Gray Level Emphasis from GLDM in LOG preprocessed image of CTD sigma value 5) for EGFR and KRAS, respectively. Furthermore, the machine learning algorithm improved the perdition power up to AUC: 0.82 for EGFR (LOG preprocessed of PET image set with sigma 3 with VT feature selector and SGD classifier) and AUC: 0.83 for KRAS (CT image set with sigma 3.5 with SM feature selector and SGD classifier). Conclusion: We demonstrated that radiomic features extracted from different image-feature sets could be used for EGFR and KRAS mutation status prediction in NSCLC patients, and showed that they have more predictive power than conventional imaging parameters.
PET/CT Radiomic Sequencer for Prediction of EGFR and KRAS Mutation Status in NSCLC Patients
Jun 15, 2019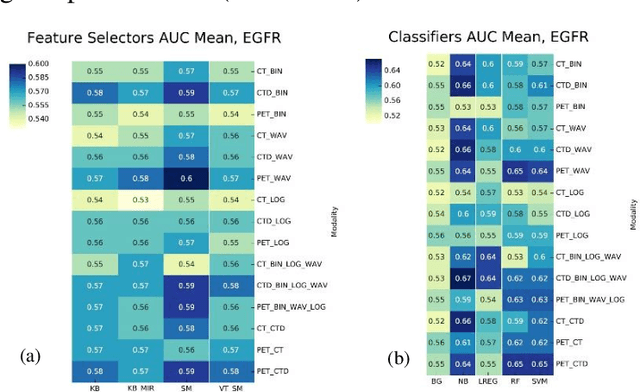
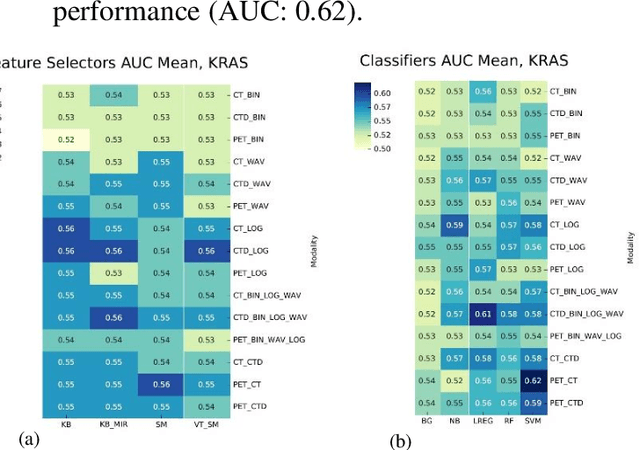
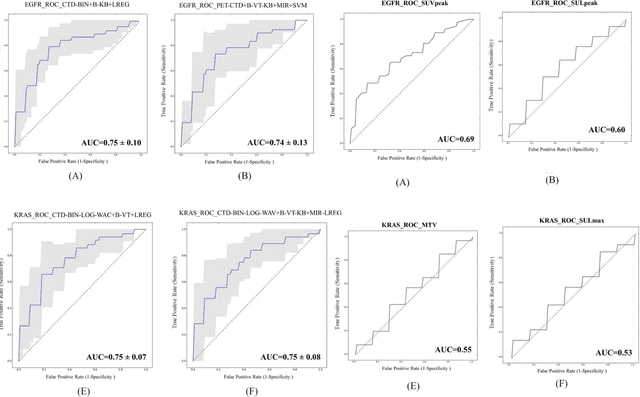
Abstract:The aim of this study was to develop radiomic models using PET/CT radiomic features with different machine learning approaches for finding best predictive epidermal growth factor receptor (EGFR) and Kirsten rat sarcoma viral oncogene (KRAS) mutation status. Patients images including PET and CT [diagnostic (CTD) and low dose CT (CTA)] were pre-processed using wavelet (WAV), Laplacian of Gaussian (LOG) and 64 bin discretization (BIN) (alone or in combinations) and several features from images were extracted. The prediction performance of model was checked using the area under the receiver operator characteristic (ROC) curve (AUC). Results showed a wide range of radiomic model AUC performances up to 0.75 in prediction of EGFR and KRAS mutation status. Combination of K-Best and variance threshold feature selector with logistic regression (LREG) classifier in diagnostic CT scan led to the best performance in EGFR (CTD-BIN+B-KB+LREG, AUC: mean 0.75 sd 0.10) and KRAS (CTD-BIN-LOG-WAV+B-VT+LREG, AUC: mean 0.75 sd 0.07) respectively. Additionally, incorporating PET, kept AUC values at ~0.74. When considering conventional features only, highest predictive performance was achieved by PET SUVpeak (AUC: 0.69) for EGFR and by PET MTV (AUC: 0.55) for KRAS. In comparison with conventional PET parameters such as standard uptake value, radiomic models were found as more predictive. Our findings demonstrated that non-invasive and reliable radiomics analysis can be successfully used to predict EGFR and KRAS mutation status in NSCLC patients.
Reliability of PET/CT shape and heterogeneity features in functional and morphological components of Non-Small Cell Lung Cancer tumors: a repeatability analysis in a prospective multi-center cohort
Oct 05, 2016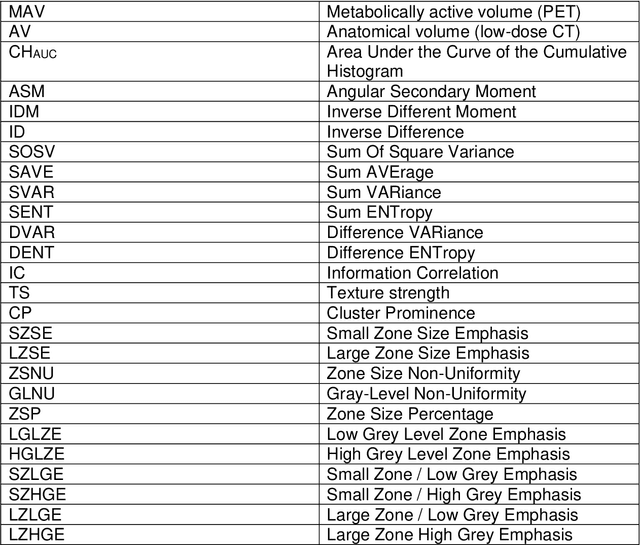
Abstract:Purpose: The main purpose of this study was to assess the reliability of shape and heterogeneity features in both Positron Emission Tomography (PET) and low-dose Computed Tomography (CT) components of PET/CT. A secondary objective was to investigate the impact of image quantization.Material and methods: A Health Insurance Portability and Accountability Act -compliant secondary analysis of deidentified prospectively acquired PET/CT test-retest datasets of 74 patients from multi-center Merck and ACRIN trials was performed. Metabolically active volumes were automatically delineated on PET with Fuzzy Locally Adaptive Bayesian algorithm. 3DSlicerTM was used to semi-automatically delineate the anatomical volumes on low-dose CT components. Two quantization methods were considered: a quantization into a set number of bins (quantizationB) and an alternative quantization with bins of fixed width (quantizationW). Four shape descriptors, ten first-order metrics and 26 textural features were computed. Bland-Altman analysis was used to quantify repeatability. Features were subsequently categorized as very reliable, reliable, moderately reliable and poorly reliable with respect to the corresponding volume variability. Results: Repeatability was highly variable amongst features. Numerous metrics were identified as poorly or moderately reliable. Others were (very) reliable in both modalities, and in all categories (shape, 1st-, 2nd- and 3rd-order metrics). Image quantization played a major role in the features repeatability. Features were more reliable in PET with quantizationB, whereas quantizationW showed better results in CT.Conclusion: The test-retest repeatability of shape and heterogeneity features in PET and low-dose CT varied greatly amongst metrics. The level of repeatability also depended strongly on the quantization step, with different optimal choices for each modality. The repeatability of PET and low-dose CT features should be carefully taken into account when selecting metrics to build multiparametric models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge