Hassan Maleki
Non-Invasive MGMT Status Prediction in GBM Cancer Using Magnetic Resonance Images Radiomics Features: Univariate and Multivariate Machine Learning Radiogenomics Analysis
Jul 08, 2019



Abstract:Background and aim: This study aimed to predict methylation status of the O-6 methyl guanine-DNA methyl transferase (MGMT) gene promoter status by using MRI radiomics features, as well as univariate and multivariate analysis. Material and Methods: Eighty-two patients who had a MGMT methylation status were include in this study. Tumor were manually segmented in the four regions of MR images, a) whole tumor, b) active/enhanced region, c) necrotic regions and d) edema regions (E). About seven thousand radiomics features were extracted for each patient. Feature selection and classifier were used to predict MGMT status through different machine learning algorithms. The area under the curve (AUC) of receiver operating characteristic (ROC) curve was used for model evaluations. Results: Regarding univariate analysis, the Inverse Variance feature from gray level co-occurrence matrix (GLCM) in Whole Tumor segment with 4.5 mm Sigma of Laplacian of Gaussian filter with AUC: 0.71 (p-value: 0.002) was found to be the best predictor. For multivariate analysis, the decision tree classifier with Select from Model feature selector and LOG filter in Edema region had the highest performance (AUC: 0.78), followed by Ada Boost classifier with Select from Model feature selector and LOG filter in Edema region (AUC: 0.74). Conclusion: This study showed that radiomics using machine learning algorithms is a feasible, noninvasive approach to predict MGMT methylation status in GBM cancer patients Keywords: Radiomics, Radiogenomics, GBM, MRI, MGMT
Next Generation Radiogenomics Sequencing for Prediction of EGFR and KRAS Mutation Status in NSCLC Patients Using Multimodal Imaging and Machine Learning Approaches
Jul 03, 2019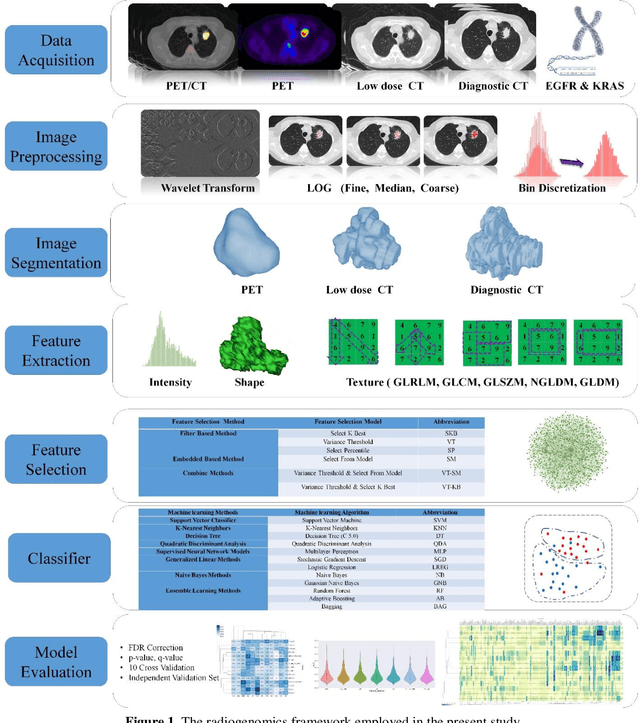
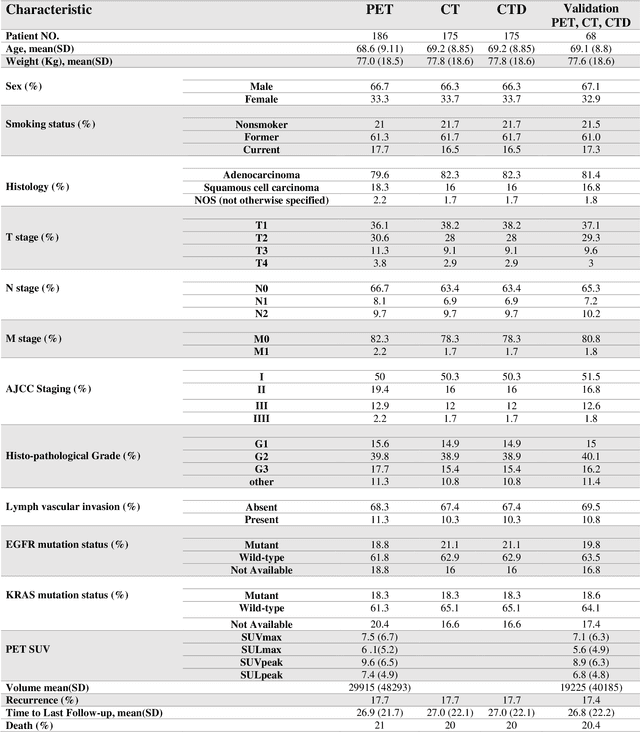

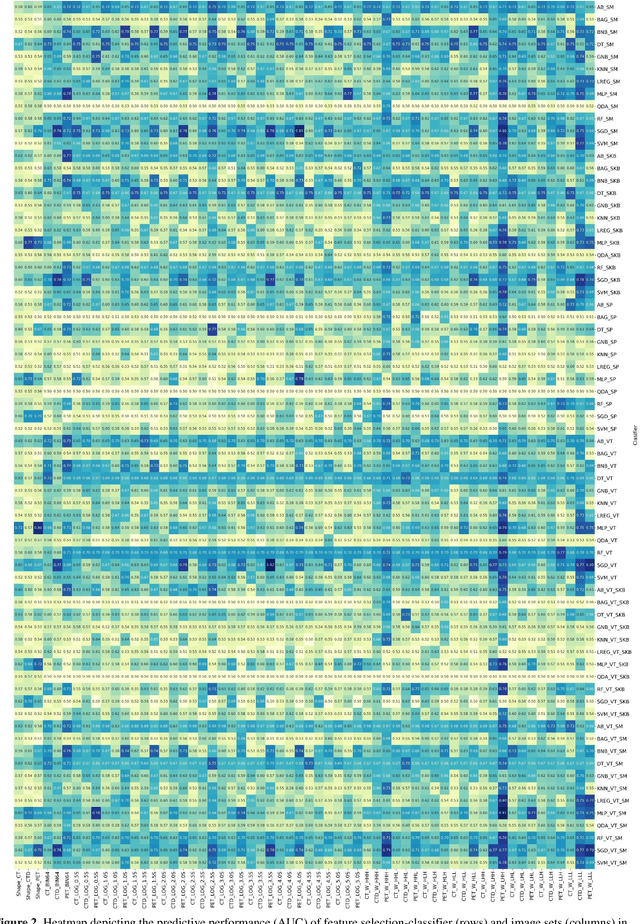
Abstract:Aim: In the present work, we aimed to evaluate a comprehensive radiomics framework that enabled prediction of EGFR and KRAS mutation status in NSCLC cancer patients based on PET and CT multi-modalities radiomic features and machine learning (ML) algorithms. Methods: Our study involved 211 NSCLC cancer patient with PET and CTD images. More than twenty thousand radiomic features from different image-feature sets were extracted Feature value was normalized to obtain Z-scores, followed by student t-test students for comparison, high correlated features were eliminated and the False discovery rate (FDR) correction were performed Six feature selection methods and twelve classifiers were used to predict gene status in patient and model evaluation was reported on independent validation sets (68 patients). Results: The best predictive power of conventional PET parameters was achieved by SUVpeak (AUC: 0.69, P-value = 0.0002) and MTV (AUC: 0.55, P-value = 0.0011) for EGFR and KRAS, respectively. Univariate analysis of radiomics features improved prediction power up to AUC: 75 (q-value: 0.003, Short Run Emphasis feature of GLRLM from LOG preprocessed image of PET with sigma value 1.5) and AUC: 0.71 (q-value 0.00005, The Large Dependence Low Gray Level Emphasis from GLDM in LOG preprocessed image of CTD sigma value 5) for EGFR and KRAS, respectively. Furthermore, the machine learning algorithm improved the perdition power up to AUC: 0.82 for EGFR (LOG preprocessed of PET image set with sigma 3 with VT feature selector and SGD classifier) and AUC: 0.83 for KRAS (CT image set with sigma 3.5 with SM feature selector and SGD classifier). Conclusion: We demonstrated that radiomic features extracted from different image-feature sets could be used for EGFR and KRAS mutation status prediction in NSCLC patients, and showed that they have more predictive power than conventional imaging parameters.
PET/CT Radiomic Sequencer for Prediction of EGFR and KRAS Mutation Status in NSCLC Patients
Jun 15, 2019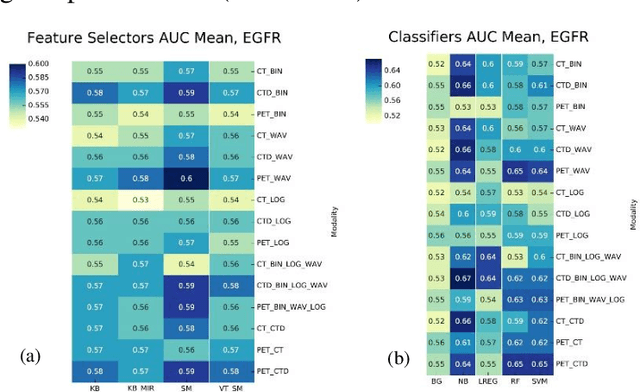
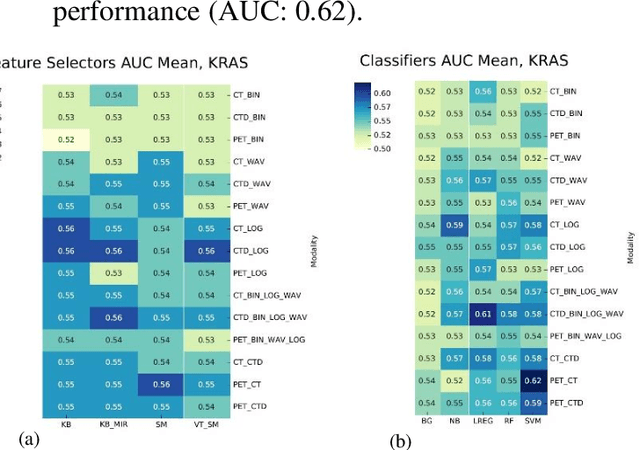
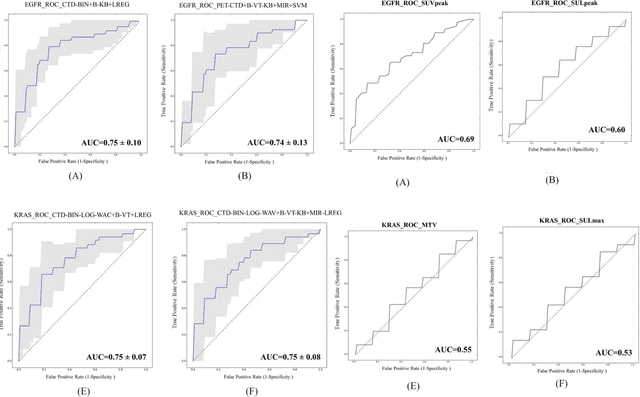
Abstract:The aim of this study was to develop radiomic models using PET/CT radiomic features with different machine learning approaches for finding best predictive epidermal growth factor receptor (EGFR) and Kirsten rat sarcoma viral oncogene (KRAS) mutation status. Patients images including PET and CT [diagnostic (CTD) and low dose CT (CTA)] were pre-processed using wavelet (WAV), Laplacian of Gaussian (LOG) and 64 bin discretization (BIN) (alone or in combinations) and several features from images were extracted. The prediction performance of model was checked using the area under the receiver operator characteristic (ROC) curve (AUC). Results showed a wide range of radiomic model AUC performances up to 0.75 in prediction of EGFR and KRAS mutation status. Combination of K-Best and variance threshold feature selector with logistic regression (LREG) classifier in diagnostic CT scan led to the best performance in EGFR (CTD-BIN+B-KB+LREG, AUC: mean 0.75 sd 0.10) and KRAS (CTD-BIN-LOG-WAV+B-VT+LREG, AUC: mean 0.75 sd 0.07) respectively. Additionally, incorporating PET, kept AUC values at ~0.74. When considering conventional features only, highest predictive performance was achieved by PET SUVpeak (AUC: 0.69) for EGFR and by PET MTV (AUC: 0.55) for KRAS. In comparison with conventional PET parameters such as standard uptake value, radiomic models were found as more predictive. Our findings demonstrated that non-invasive and reliable radiomics analysis can be successfully used to predict EGFR and KRAS mutation status in NSCLC patients.
SentiPers: A Sentiment Analysis Corpus for Persian
Jan 23, 2018


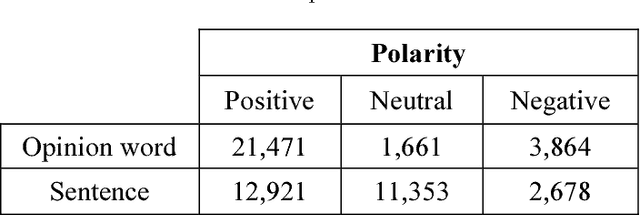
Abstract:Sentiment Analysis (SA) is a major field of study in natural language processing, computational linguistics and information retrieval. Interest in SA has been constantly growing in both academia and industry over the recent years. Moreover, there is an increasing need for generating appropriate resources and datasets in particular for low resource languages including Persian. These datasets play an important role in designing and developing appropriate opinion mining platforms using supervised, semi-supervised or unsupervised methods. In this paper, we outline the entire process of developing a manually annotated sentiment corpus, SentiPers, which covers formal and informal written contemporary Persian. To the best of our knowledge, SentiPers is a unique sentiment corpus with such a rich annotation in three different levels including document-level, sentence-level, and entity/aspect-level for Persian. The corpus contains more than 26000 sentences of users opinions from digital product domain and benefits from special characteristics such as quantifying the positiveness or negativity of an opinion through assigning a number within a specific range to any given sentence. Furthermore, we present statistics on various components of our corpus as well as studying the inter-annotator agreement among the annotators. Finally, some of the challenges that we faced during the annotation process will be discussed as well.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge