Yihao Li
Patch Progression Masked Autoencoder with Fusion CNN Network for Classifying Evolution Between Two Pairs of 2D OCT Slices
Aug 27, 2025Abstract:Age-related Macular Degeneration (AMD) is a prevalent eye condition affecting visual acuity. Anti-vascular endothelial growth factor (anti-VEGF) treatments have been effective in slowing the progression of neovascular AMD, with better outcomes achieved through timely diagnosis and consistent monitoring. Tracking the progression of neovascular activity in OCT scans of patients with exudative AMD allows for the development of more personalized and effective treatment plans. This was the focus of the Monitoring Age-related Macular Degeneration Progression in Optical Coherence Tomography (MARIO) challenge, in which we participated. In Task 1, which involved classifying the evolution between two pairs of 2D slices from consecutive OCT acquisitions, we employed a fusion CNN network with model ensembling to further enhance the model's performance. For Task 2, which focused on predicting progression over the next three months based on current exam data, we proposed the Patch Progression Masked Autoencoder that generates an OCT for the next exam and then classifies the evolution between the current OCT and the one generated using our solution from Task 1. The results we achieved allowed us to place in the Top 10 for both tasks. Some team members are part of the same organization as the challenge organizers; therefore, we are not eligible to compete for the prize.
* 10 pages, 5 figures, 3 tables, challenge/conference paper
USIS16K: High-Quality Dataset for Underwater Salient Instance Segmentation
Jun 24, 2025Abstract:Inspired by the biological visual system that selectively allocates attention to efficiently identify salient objects or regions, underwater salient instance segmentation (USIS) aims to jointly address the problems of where to look (saliency prediction) and what is there (instance segmentation) in underwater scenarios. However, USIS remains an underexplored challenge due to the inaccessibility and dynamic nature of underwater environments, as well as the scarcity of large-scale, high-quality annotated datasets. In this paper, we introduce USIS16K, a large-scale dataset comprising 16,151 high-resolution underwater images collected from diverse environmental settings and covering 158 categories of underwater objects. Each image is annotated with high-quality instance-level salient object masks, representing a significant advance in terms of diversity, complexity, and scalability. Furthermore, we provide benchmark evaluations on underwater object detection and USIS tasks using USIS16K. To facilitate future research in this domain, the dataset and benchmark models are publicly available.
SpikeStereoNet: A Brain-Inspired Framework for Stereo Depth Estimation from Spike Streams
May 26, 2025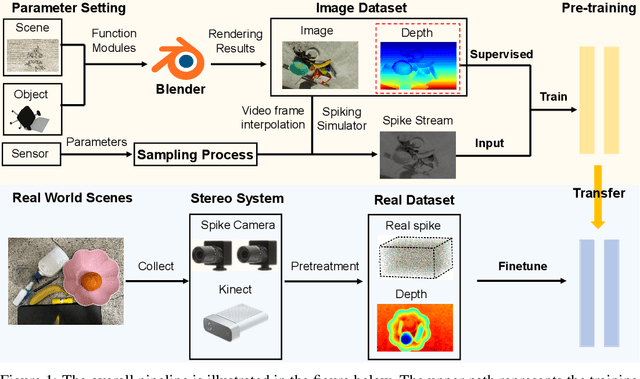
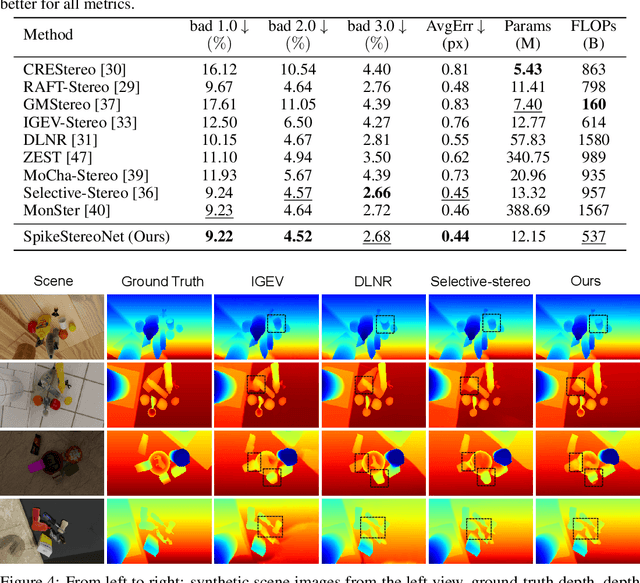
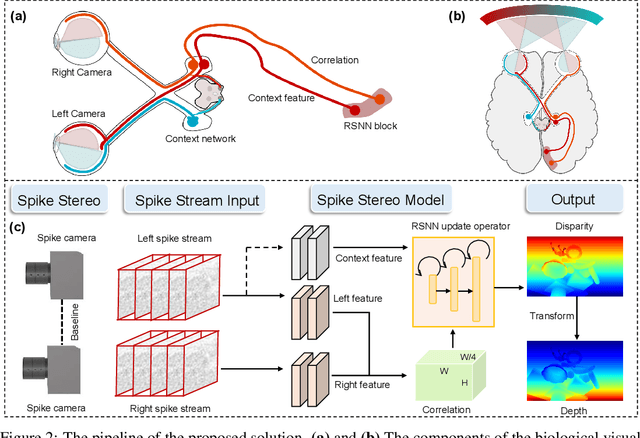
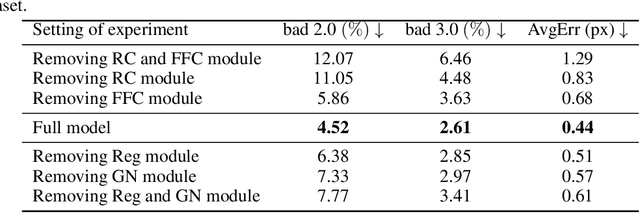
Abstract:Conventional frame-based cameras often struggle with stereo depth estimation in rapidly changing scenes. In contrast, bio-inspired spike cameras emit asynchronous events at microsecond-level resolution, providing an alternative sensing modality. However, existing methods lack specialized stereo algorithms and benchmarks tailored to the spike data. To address this gap, we propose SpikeStereoNet, a brain-inspired framework and the first to estimate stereo depth directly from raw spike streams. The model fuses raw spike streams from two viewpoints and iteratively refines depth estimation through a recurrent spiking neural network (RSNN) update module. To benchmark our approach, we introduce a large-scale synthetic spike stream dataset and a real-world stereo spike dataset with dense depth annotations. SpikeStereoNet outperforms existing methods on both datasets by leveraging spike streams' ability to capture subtle edges and intensity shifts in challenging regions such as textureless surfaces and extreme lighting conditions. Furthermore, our framework exhibits strong data efficiency, maintaining high accuracy even with substantially reduced training data. The source code and datasets will be publicly available.
UAV Cognitive Semantic Communications Enabled by Knowledge Graph for Robust Object Detection
Feb 06, 2025



Abstract:Unmanned aerial vehicles (UAVs) are widely used for object detection. However, the existing UAV-based object detection systems are subject to severe challenges, namely, their limited computation, energy and communication resources, which limits the achievable detection performance. To overcome these challenges, a UAV cognitive semantic communication system is proposed by exploiting a knowledge graph. Moreover, we design a multi-scale codec for semantic compression to reduce data transmission volume while guaranteeing detection performance. Considering the complexity and dynamicity of UAV communication scenarios, a signal-to-noise ratio (SNR) adaptive module with robust channel adaptation capability is introduced. Furthermore, an object detection scheme is proposed by exploiting the knowledge graph to overcome channel noise interference and compression distortion. Simulation results conducted on the practical aerial image dataset demonstrate that our proposed semantic communication system outperforms benchmark systems in terms of detection accuracy, communication robustness, and computation efficiency, especially in dealing with low bandwidth compression ratios and low SNR regimes.
From Intention To Implementation: Automating Biomedical Research via LLMs
Dec 12, 2024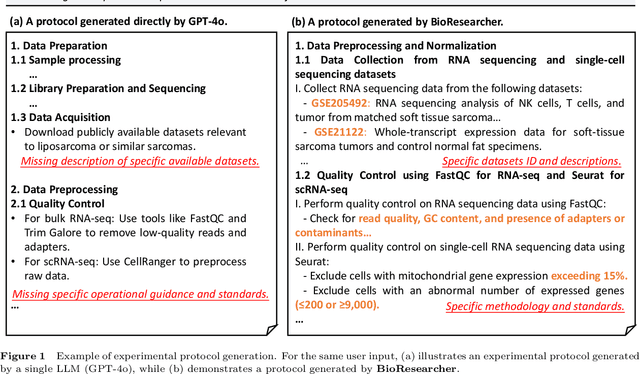
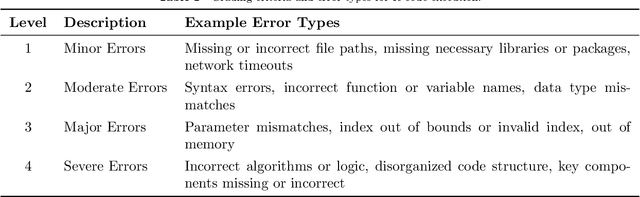
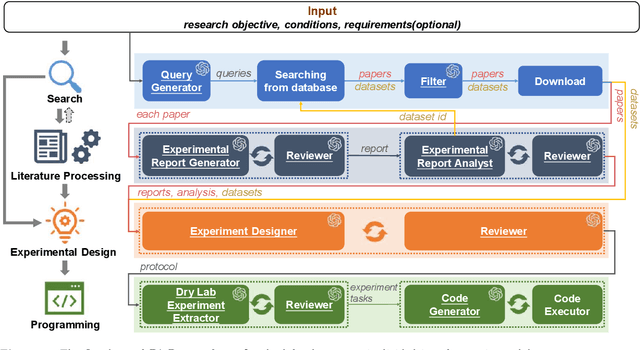
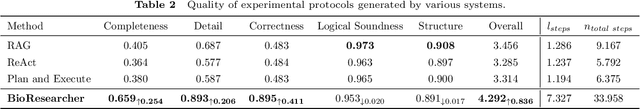
Abstract:Conventional biomedical research is increasingly labor-intensive due to the exponential growth of scientific literature and datasets. Artificial intelligence (AI), particularly Large Language Models (LLMs), has the potential to revolutionize this process by automating various steps. Still, significant challenges remain, including the need for multidisciplinary expertise, logicality of experimental design, and performance measurements. This paper introduces BioResearcher, the first end-to-end automated system designed to streamline the entire biomedical research process involving dry lab experiments. BioResearcher employs a modular multi-agent architecture, integrating specialized agents for search, literature processing, experimental design, and programming. By decomposing complex tasks into logically related sub-tasks and utilizing a hierarchical learning approach, BioResearcher effectively addresses the challenges of multidisciplinary requirements and logical complexity. Furthermore, BioResearcher incorporates an LLM-based reviewer for in-process quality control and introduces novel evaluation metrics to assess the quality and automation of experimental protocols. BioResearcher successfully achieves an average execution success rate of 63.07% across eight previously unmet research objectives. The generated protocols averagely outperform typical agent systems by 22.0% on five quality metrics. The system demonstrates significant potential to reduce researchers' workloads and accelerate biomedical discoveries, paving the way for future innovations in automated research systems.
GaussReg: Fast 3D Registration with Gaussian Splatting
Jul 07, 2024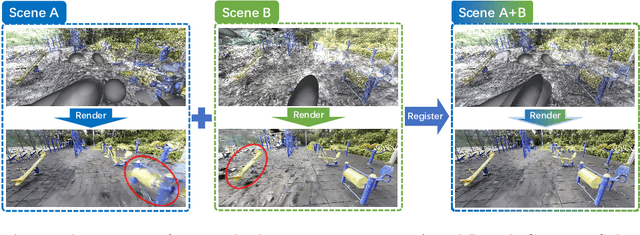
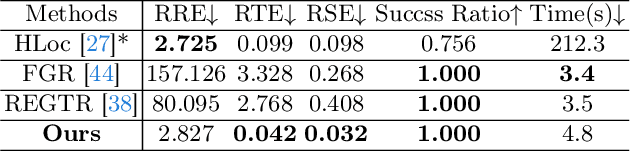
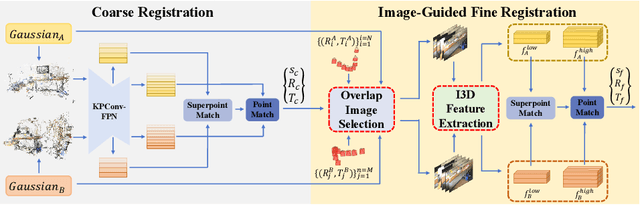
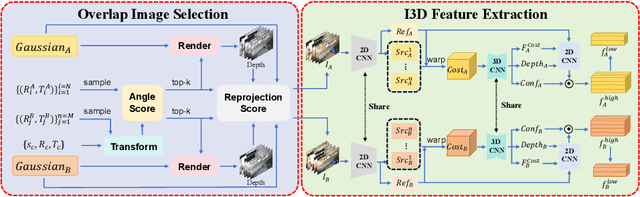
Abstract:Point cloud registration is a fundamental problem for large-scale 3D scene scanning and reconstruction. With the help of deep learning, registration methods have evolved significantly, reaching a nearly-mature stage. As the introduction of Neural Radiance Fields (NeRF), it has become the most popular 3D scene representation as its powerful view synthesis capabilities. Regarding NeRF representation, its registration is also required for large-scale scene reconstruction. However, this topic extremly lacks exploration. This is due to the inherent challenge to model the geometric relationship among two scenes with implicit representations. The existing methods usually convert the implicit representation to explicit representation for further registration. Most recently, Gaussian Splatting (GS) is introduced, employing explicit 3D Gaussian. This method significantly enhances rendering speed while maintaining high rendering quality. Given two scenes with explicit GS representations, in this work, we explore the 3D registration task between them. To this end, we propose GaussReg, a novel coarse-to-fine framework, both fast and accurate. The coarse stage follows existing point cloud registration methods and estimates a rough alignment for point clouds from GS. We further newly present an image-guided fine registration approach, which renders images from GS to provide more detailed geometric information for precise alignment. To support comprehensive evaluation, we carefully build a scene-level dataset called ScanNet-GSReg with 1379 scenes obtained from the ScanNet dataset and collect an in-the-wild dataset called GSReg. Experimental results demonstrate our method achieves state-of-the-art performance on multiple datasets. Our GaussReg is 44 times faster than HLoc (SuperPoint as the feature extractor and SuperGlue as the matcher) with comparable accuracy.
LaTiM: Longitudinal representation learning in continuous-time models to predict disease progression
Apr 10, 2024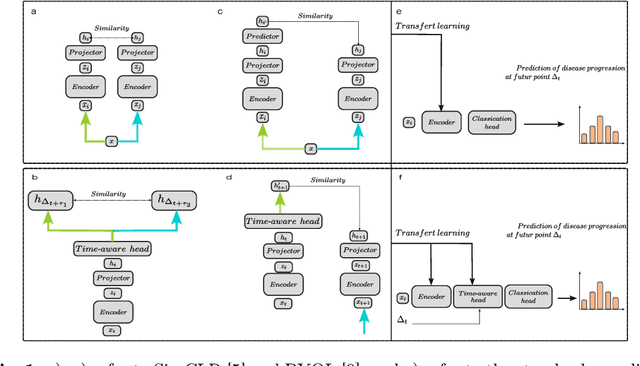
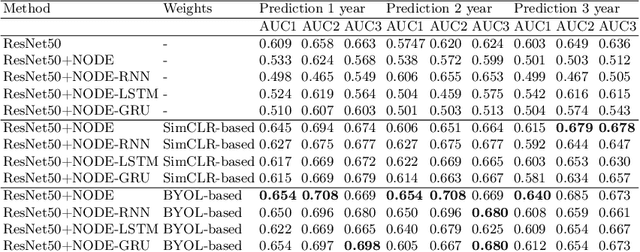
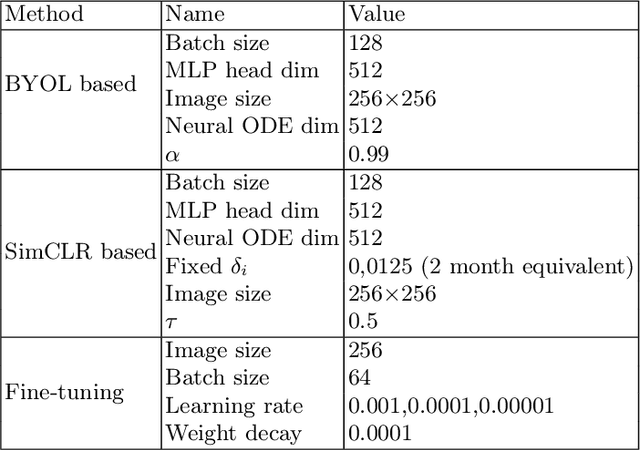
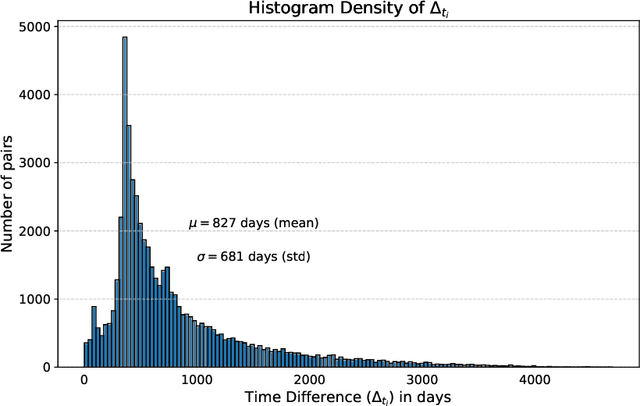
Abstract:This work proposes a novel framework for analyzing disease progression using time-aware neural ordinary differential equations (NODE). We introduce a "time-aware head" in a framework trained through self-supervised learning (SSL) to leverage temporal information in latent space for data augmentation. This approach effectively integrates NODEs with SSL, offering significant performance improvements compared to traditional methods that lack explicit temporal integration. We demonstrate the effectiveness of our strategy for diabetic retinopathy progression prediction using the OPHDIAT database. Compared to the baseline, all NODE architectures achieve statistically significant improvements in area under the ROC curve (AUC) and Kappa metrics, highlighting the efficacy of pre-training with SSL-inspired approaches. Additionally, our framework promotes stable training for NODEs, a commonly encountered challenge in time-aware modeling.
L-MAE: Longitudinal masked auto-encoder with time and severity-aware encoding for diabetic retinopathy progression prediction
Mar 24, 2024Abstract:Pre-training strategies based on self-supervised learning (SSL) have proven to be effective pretext tasks for many downstream tasks in computer vision. Due to the significant disparity between medical and natural images, the application of typical SSL is not straightforward in medical imaging. Additionally, those pretext tasks often lack context, which is critical for computer-aided clinical decision support. In this paper, we developed a longitudinal masked auto-encoder (MAE) based on the well-known Transformer-based MAE. In particular, we explored the importance of time-aware position embedding as well as disease progression-aware masking. Taking into account the time between examinations instead of just scheduling them offers the benefit of capturing temporal changes and trends. The masking strategy, for its part, evolves during follow-up to better capture pathological changes, ensuring a more accurate assessment of disease progression. Using OPHDIAT, a large follow-up screening dataset targeting diabetic retinopathy (DR), we evaluated the pre-trained weights on a longitudinal task, which is to predict the severity label of the next visit within 3 years based on the past time series examinations. Our results demonstrated the relevancy of both time-aware position embedding and masking strategies based on disease progression knowledge. Compared to popular baseline models and standard longitudinal Transformers, these simple yet effective extensions significantly enhance the predictive ability of deep classification models.
An Enhanced Prompt-Based LLM Reasoning Scheme via Knowledge Graph-Integrated Collaboration
Feb 07, 2024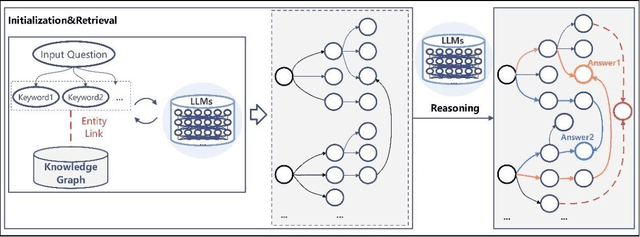
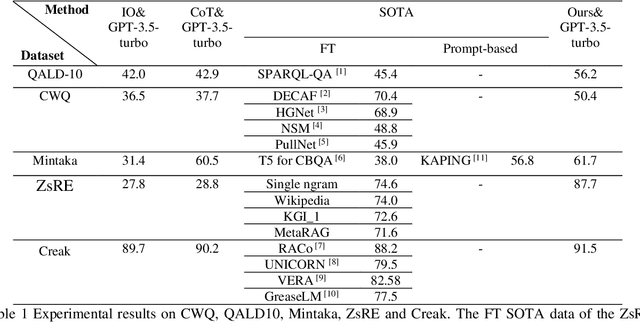
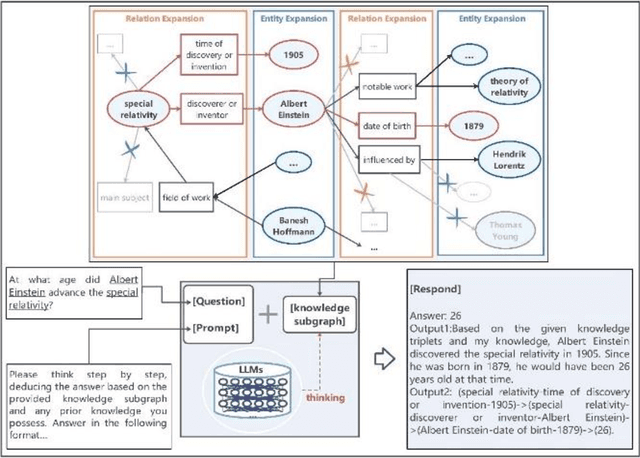

Abstract:While Large Language Models (LLMs) demonstrate exceptional performance in a multitude of Natural Language Processing (NLP) tasks, they encounter challenges in practical applications, including issues with hallucinations, inadequate knowledge updating, and limited transparency in the reasoning process. To overcome these limitations, this study innovatively proposes a collaborative training-free reasoning scheme involving tight cooperation between Knowledge Graph (KG) and LLMs. This scheme first involves using LLMs to iteratively explore KG, selectively retrieving a task-relevant knowledge subgraph to support reasoning. The LLMs are then guided to further combine inherent implicit knowledge to reason on the subgraph while explicitly elucidating the reasoning process. Through such a cooperative approach, our scheme achieves more reliable knowledge-based reasoning and facilitates the tracing of the reasoning results. Experimental results show that our scheme significantly progressed across multiple datasets, notably achieving over a 10% improvement on the QALD10 dataset compared to the best baseline and the fine-tuned state-of-the-art (SOTA) work. Building on this success, this study hopes to offer a valuable reference for future research in the fusion of KG and LLMs, thereby enhancing LLMs' proficiency in solving complex issues.
DISCOVER: 2-D Multiview Summarization of Optical Coherence Tomography Angiography for Automatic Diabetic Retinopathy Diagnosis
Jan 10, 2024Abstract:Diabetic Retinopathy (DR), an ocular complication of diabetes, is a leading cause of blindness worldwide. Traditionally, DR is monitored using Color Fundus Photography (CFP), a widespread 2-D imaging modality. However, DR classifications based on CFP have poor predictive power, resulting in suboptimal DR management. Optical Coherence Tomography Angiography (OCTA) is a recent 3-D imaging modality offering enhanced structural and functional information (blood flow) with a wider field of view. This paper investigates automatic DR severity assessment using 3-D OCTA. A straightforward solution to this task is a 3-D neural network classifier. However, 3-D architectures have numerous parameters and typically require many training samples. A lighter solution consists in using 2-D neural network classifiers processing 2-D en-face (or frontal) projections and/or 2-D cross-sectional slices. Such an approach mimics the way ophthalmologists analyze OCTA acquisitions: 1) en-face flow maps are often used to detect avascular zones and neovascularization, and 2) cross-sectional slices are commonly analyzed to detect macular edemas, for instance. However, arbitrary data reduction or selection might result in information loss. Two complementary strategies are thus proposed to optimally summarize OCTA volumes with 2-D images: 1) a parametric en-face projection optimized through deep learning and 2) a cross-sectional slice selection process controlled through gradient-based attribution. The full summarization and DR classification pipeline is trained from end to end. The automatic 2-D summary can be displayed in a viewer or printed in a report to support the decision. We show that the proposed 2-D summarization and classification pipeline outperforms direct 3-D classification with the advantage of improved interpretability.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge