Pierre-Henri Conze
Deep learning-enabled prediction of surgical errors during cataract surgery: from simulation to real-world application
Mar 28, 2025Abstract:Real-time prediction of technical errors from cataract surgical videos can be highly beneficial, particularly for telementoring, which involves remote guidance and mentoring through digital platforms. However, the rarity of surgical errors makes their detection and analysis challenging using artificial intelligence. To tackle this issue, we leveraged videos from the EyeSi Surgical cataract surgery simulator to learn to predict errors and transfer the acquired knowledge to real-world surgical contexts. By employing deep learning models, we demonstrated the feasibility of making real-time predictions using simulator data with a very short temporal history, enabling on-the-fly computations. We then transferred these insights to real-world settings through unsupervised domain adaptation, without relying on labeled videos from real surgeries for training, which are limited. This was achieved by aligning video clips from the simulator with real-world footage and pre-training the models using pretext tasks on both simulated and real surgical data. For a 1-second prediction window on the simulator, we achieved an overall AUC of 0.820 for error prediction using 600$\times$600 pixel images, and 0.784 using smaller 299$\times$299 pixel images. In real-world settings, we obtained an AUC of up to 0.663 with domain adaptation, marking an improvement over direct model application without adaptation, which yielded an AUC of 0.578. To our knowledge, this is the first work to address the tasks of learning surgical error prediction on a simulator using video data only and transferring this knowledge to real-world cataract surgery.
LaTiM: Longitudinal representation learning in continuous-time models to predict disease progression
Apr 10, 2024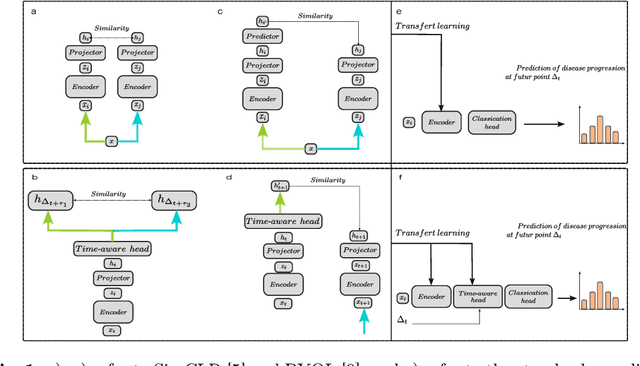
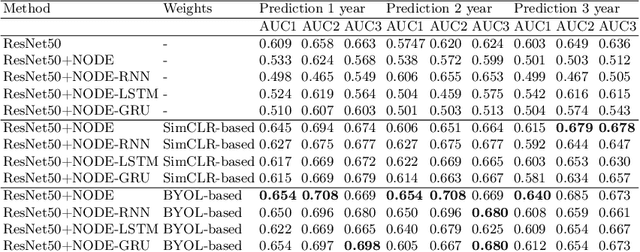
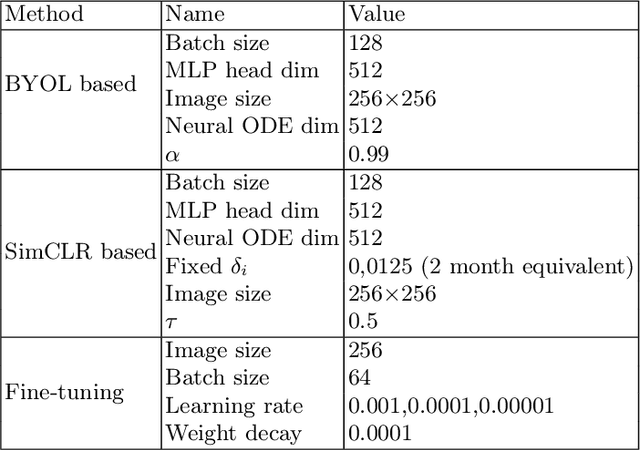
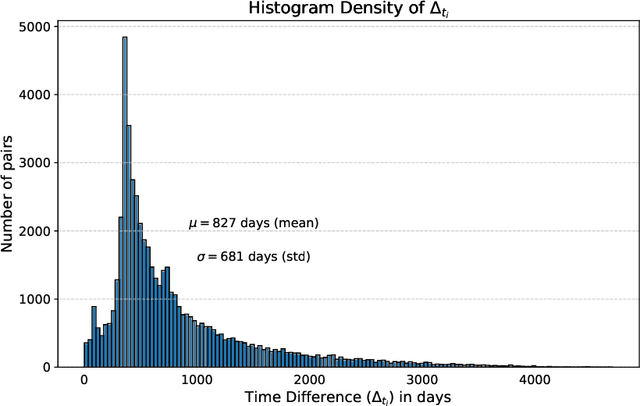
Abstract:This work proposes a novel framework for analyzing disease progression using time-aware neural ordinary differential equations (NODE). We introduce a "time-aware head" in a framework trained through self-supervised learning (SSL) to leverage temporal information in latent space for data augmentation. This approach effectively integrates NODEs with SSL, offering significant performance improvements compared to traditional methods that lack explicit temporal integration. We demonstrate the effectiveness of our strategy for diabetic retinopathy progression prediction using the OPHDIAT database. Compared to the baseline, all NODE architectures achieve statistically significant improvements in area under the ROC curve (AUC) and Kappa metrics, highlighting the efficacy of pre-training with SSL-inspired approaches. Additionally, our framework promotes stable training for NODEs, a commonly encountered challenge in time-aware modeling.
Guidelines for Cerebrovascular Segmentation: Managing Imperfect Annotations in the context of Semi-Supervised Learning
Apr 02, 2024Abstract:Segmentation in medical imaging is an essential and often preliminary task in the image processing chain, driving numerous efforts towards the design of robust segmentation algorithms. Supervised learning methods achieve excellent performances when fed with a sufficient amount of labeled data. However, such labels are typically highly time-consuming, error-prone and expensive to produce. Alternatively, semi-supervised learning approaches leverage both labeled and unlabeled data, and are very useful when only a small fraction of the dataset is labeled. They are particularly useful for cerebrovascular segmentation, given that labeling a single volume requires several hours for an expert. In addition to the challenge posed by insufficient annotations, there are concerns regarding annotation consistency. The task of annotating the cerebrovascular tree is inherently ambiguous. Due to the discrete nature of images, the borders and extremities of vessels are often unclear. Consequently, annotations heavily rely on the expert subjectivity and on the underlying clinical objective. These discrepancies significantly increase the complexity of the segmentation task for the model and consequently impair the results. Consequently, it becomes imperative to provide clinicians with precise guidelines to improve the annotation process and construct more uniform datasets. In this article, we investigate the data dependency of deep learning methods within the context of imperfect data and semi-supervised learning, for cerebrovascular segmentation. Specifically, this study compares various state-of-the-art semi-supervised methods based on unsupervised regularization and evaluates their performance in diverse quantity and quality data scenarios. Based on these experiments, we provide guidelines for the annotation and training of cerebrovascular segmentation models.
L-MAE: Longitudinal masked auto-encoder with time and severity-aware encoding for diabetic retinopathy progression prediction
Mar 24, 2024Abstract:Pre-training strategies based on self-supervised learning (SSL) have proven to be effective pretext tasks for many downstream tasks in computer vision. Due to the significant disparity between medical and natural images, the application of typical SSL is not straightforward in medical imaging. Additionally, those pretext tasks often lack context, which is critical for computer-aided clinical decision support. In this paper, we developed a longitudinal masked auto-encoder (MAE) based on the well-known Transformer-based MAE. In particular, we explored the importance of time-aware position embedding as well as disease progression-aware masking. Taking into account the time between examinations instead of just scheduling them offers the benefit of capturing temporal changes and trends. The masking strategy, for its part, evolves during follow-up to better capture pathological changes, ensuring a more accurate assessment of disease progression. Using OPHDIAT, a large follow-up screening dataset targeting diabetic retinopathy (DR), we evaluated the pre-trained weights on a longitudinal task, which is to predict the severity label of the next visit within 3 years based on the past time series examinations. Our results demonstrated the relevancy of both time-aware position embedding and masking strategies based on disease progression knowledge. Compared to popular baseline models and standard longitudinal Transformers, these simple yet effective extensions significantly enhance the predictive ability of deep classification models.
DISCOVER: 2-D Multiview Summarization of Optical Coherence Tomography Angiography for Automatic Diabetic Retinopathy Diagnosis
Jan 10, 2024Abstract:Diabetic Retinopathy (DR), an ocular complication of diabetes, is a leading cause of blindness worldwide. Traditionally, DR is monitored using Color Fundus Photography (CFP), a widespread 2-D imaging modality. However, DR classifications based on CFP have poor predictive power, resulting in suboptimal DR management. Optical Coherence Tomography Angiography (OCTA) is a recent 3-D imaging modality offering enhanced structural and functional information (blood flow) with a wider field of view. This paper investigates automatic DR severity assessment using 3-D OCTA. A straightforward solution to this task is a 3-D neural network classifier. However, 3-D architectures have numerous parameters and typically require many training samples. A lighter solution consists in using 2-D neural network classifiers processing 2-D en-face (or frontal) projections and/or 2-D cross-sectional slices. Such an approach mimics the way ophthalmologists analyze OCTA acquisitions: 1) en-face flow maps are often used to detect avascular zones and neovascularization, and 2) cross-sectional slices are commonly analyzed to detect macular edemas, for instance. However, arbitrary data reduction or selection might result in information loss. Two complementary strategies are thus proposed to optimally summarize OCTA volumes with 2-D images: 1) a parametric en-face projection optimized through deep learning and 2) a cross-sectional slice selection process controlled through gradient-based attribution. The full summarization and DR classification pipeline is trained from end to end. The automatic 2-D summary can be displayed in a viewer or printed in a report to support the decision. We show that the proposed 2-D summarization and classification pipeline outperforms direct 3-D classification with the advantage of improved interpretability.
Automated Detection of Myopic Maculopathy in MMAC 2023: Achievements in Classification, Segmentation, and Spherical Equivalent Prediction
Jan 08, 2024Abstract:Myopic macular degeneration is the most common complication of myopia and the primary cause of vision loss in individuals with pathological myopia. Early detection and prompt treatment are crucial in preventing vision impairment due to myopic maculopathy. This was the focus of the Myopic Maculopathy Analysis Challenge (MMAC), in which we participated. In task 1, classification of myopic maculopathy, we employed the contrastive learning framework, specifically SimCLR, to enhance classification accuracy by effectively capturing enriched features from unlabeled data. This approach not only improved the intrinsic understanding of the data but also elevated the performance of our classification model. For Task 2 (segmentation of myopic maculopathy plus lesions), we have developed independent segmentation models tailored for different lesion segmentation tasks and implemented a test-time augmentation strategy to further enhance the model's performance. As for Task 3 (prediction of spherical equivalent), we have designed a deep regression model based on the data distribution of the dataset and employed an integration strategy to enhance the model's prediction accuracy. The results we obtained are promising and have allowed us to position ourselves in the Top 6 of the classification task, the Top 2 of the segmentation task, and the Top 1 of the prediction task. The code is available at \url{https://github.com/liyihao76/MMAC_LaTIM_Solution}.
LMT: Longitudinal Mixing Training, a Framework to Predict Disease Progression from a Single Image
Oct 16, 2023Abstract:Longitudinal imaging is able to capture both static anatomical structures and dynamic changes in disease progression toward earlier and better patient-specific pathology management. However, conventional approaches rarely take advantage of longitudinal information for detection and prediction purposes, especially for Diabetic Retinopathy (DR). In the past years, Mix-up training and pretext tasks with longitudinal context have effectively enhanced DR classification results and captured disease progression. In the meantime, a novel type of neural network named Neural Ordinary Differential Equation (NODE) has been proposed for solving ordinary differential equations, with a neural network treated as a black box. By definition, NODE is well suited for solving time-related problems. In this paper, we propose to combine these three aspects to detect and predict DR progression. Our framework, Longitudinal Mixing Training (LMT), can be considered both as a regularizer and as a pretext task that encodes the disease progression in the latent space. Additionally, we evaluate the trained model weights on a downstream task with a longitudinal context using standard and longitudinal pretext tasks. We introduce a new way to train time-aware models using $t_{mix}$, a weighted average time between two consecutive examinations. We compare our approach to standard mixing training on DR classification using OPHDIAT a longitudinal retinal Color Fundus Photographs (CFP) dataset. We were able to predict whether an eye would develop a severe DR in the following visit using a single image, with an AUC of 0.798 compared to baseline results of 0.641. Our results indicate that our longitudinal pretext task can learn the progression of DR disease and that introducing $t_{mix}$ augmentation is beneficial for time-aware models.
Longitudinal Self-supervised Learning Using Neural Ordinary Differential Equation
Oct 16, 2023Abstract:Longitudinal analysis in medical imaging is crucial to investigate the progressive changes in anatomical structures or disease progression over time. In recent years, a novel class of algorithms has emerged with the goal of learning disease progression in a self-supervised manner, using either pairs of consecutive images or time series of images. By capturing temporal patterns without external labels or supervision, longitudinal self-supervised learning (LSSL) has become a promising avenue. To better understand this core method, we explore in this paper the LSSL algorithm under different scenarios. The original LSSL is embedded in an auto-encoder (AE) structure. However, conventional self-supervised strategies are usually implemented in a Siamese-like manner. Therefore, (as a first novelty) in this study, we explore the use of Siamese-like LSSL. Another new core framework named neural ordinary differential equation (NODE). NODE is a neural network architecture that learns the dynamics of ordinary differential equations (ODE) through the use of neural networks. Many temporal systems can be described by ODE, including modeling disease progression. We believe that there is an interesting connection to make between LSSL and NODE. This paper aims at providing a better understanding of those core algorithms for learning the disease progression with the mentioned change. In our different experiments, we employ a longitudinal dataset, named OPHDIAT, targeting diabetic retinopathy (DR) follow-up. Our results demonstrate the application of LSSL without including a reconstruction term, as well as the potential of incorporating NODE in conjunction with LSSL.
Improved Automatic Diabetic Retinopathy Severity Classification Using Deep Multimodal Fusion of UWF-CFP and OCTA Images
Oct 03, 2023Abstract:Diabetic Retinopathy (DR), a prevalent and severe complication of diabetes, affects millions of individuals globally, underscoring the need for accurate and timely diagnosis. Recent advancements in imaging technologies, such as Ultra-WideField Color Fundus Photography (UWF-CFP) imaging and Optical Coherence Tomography Angiography (OCTA), provide opportunities for the early detection of DR but also pose significant challenges given the disparate nature of the data they produce. This study introduces a novel multimodal approach that leverages these imaging modalities to notably enhance DR classification. Our approach integrates 2D UWF-CFP images and 3D high-resolution 6x6 mm$^3$ OCTA (both structure and flow) images using a fusion of ResNet50 and 3D-ResNet50 models, with Squeeze-and-Excitation (SE) blocks to amplify relevant features. Additionally, to increase the model's generalization capabilities, a multimodal extension of Manifold Mixup, applied to concatenated multimodal features, is implemented. Experimental results demonstrate a remarkable enhancement in DR classification performance with the proposed multimodal approach compared to methods relying on a single modality only. The methodology laid out in this work holds substantial promise for facilitating more accurate, early detection of DR, potentially improving clinical outcomes for patients.
Cross-dimensional transfer learning in medical image segmentation with deep learning
Jul 29, 2023Abstract:Over the last decade, convolutional neural networks have emerged and advanced the state-of-the-art in various image analysis and computer vision applications. The performance of 2D image classification networks is constantly improving and being trained on databases made of millions of natural images. However, progress in medical image analysis has been hindered by limited annotated data and acquisition constraints. These limitations are even more pronounced given the volumetry of medical imaging data. In this paper, we introduce an efficient way to transfer the efficiency of a 2D classification network trained on natural images to 2D, 3D uni- and multi-modal medical image segmentation applications. In this direction, we designed novel architectures based on two key principles: weight transfer by embedding a 2D pre-trained encoder into a higher dimensional U-Net, and dimensional transfer by expanding a 2D segmentation network into a higher dimension one. The proposed networks were tested on benchmarks comprising different modalities: MR, CT, and ultrasound images. Our 2D network ranked first on the CAMUS challenge dedicated to echo-cardiographic data segmentation and surpassed the state-of-the-art. Regarding 2D/3D MR and CT abdominal images from the CHAOS challenge, our approach largely outperformed the other 2D-based methods described in the challenge paper on Dice, RAVD, ASSD, and MSSD scores and ranked third on the online evaluation platform. Our 3D network applied to the BraTS 2022 competition also achieved promising results, reaching an average Dice score of 91.69% (91.22%) for the whole tumor, 83.23% (84.77%) for the tumor core, and 81.75% (83.88%) for enhanced tumor using the approach based on weight (dimensional) transfer. Experimental and qualitative results illustrate the effectiveness of our methods for multi-dimensional medical image segmentation.
* 30 pages, 12 figures, 6 tables, Accepted for publication in the Journal of Medical Image Analysis
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge