Pascal Massin
LaTiM: Longitudinal representation learning in continuous-time models to predict disease progression
Apr 10, 2024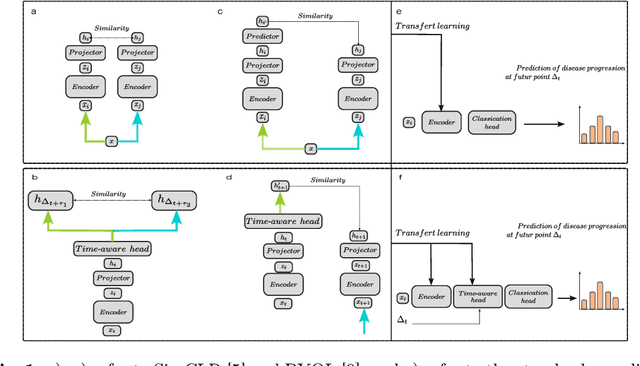
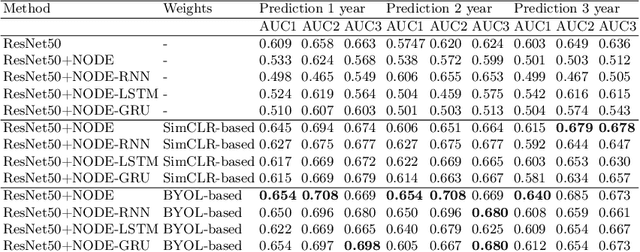
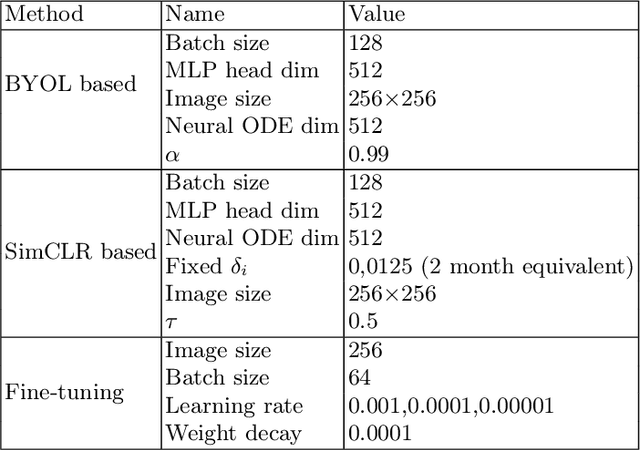
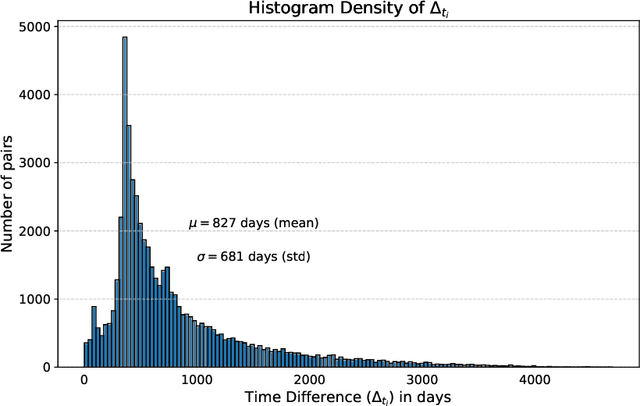
Abstract:This work proposes a novel framework for analyzing disease progression using time-aware neural ordinary differential equations (NODE). We introduce a "time-aware head" in a framework trained through self-supervised learning (SSL) to leverage temporal information in latent space for data augmentation. This approach effectively integrates NODEs with SSL, offering significant performance improvements compared to traditional methods that lack explicit temporal integration. We demonstrate the effectiveness of our strategy for diabetic retinopathy progression prediction using the OPHDIAT database. Compared to the baseline, all NODE architectures achieve statistically significant improvements in area under the ROC curve (AUC) and Kappa metrics, highlighting the efficacy of pre-training with SSL-inspired approaches. Additionally, our framework promotes stable training for NODEs, a commonly encountered challenge in time-aware modeling.
L-MAE: Longitudinal masked auto-encoder with time and severity-aware encoding for diabetic retinopathy progression prediction
Mar 24, 2024Abstract:Pre-training strategies based on self-supervised learning (SSL) have proven to be effective pretext tasks for many downstream tasks in computer vision. Due to the significant disparity between medical and natural images, the application of typical SSL is not straightforward in medical imaging. Additionally, those pretext tasks often lack context, which is critical for computer-aided clinical decision support. In this paper, we developed a longitudinal masked auto-encoder (MAE) based on the well-known Transformer-based MAE. In particular, we explored the importance of time-aware position embedding as well as disease progression-aware masking. Taking into account the time between examinations instead of just scheduling them offers the benefit of capturing temporal changes and trends. The masking strategy, for its part, evolves during follow-up to better capture pathological changes, ensuring a more accurate assessment of disease progression. Using OPHDIAT, a large follow-up screening dataset targeting diabetic retinopathy (DR), we evaluated the pre-trained weights on a longitudinal task, which is to predict the severity label of the next visit within 3 years based on the past time series examinations. Our results demonstrated the relevancy of both time-aware position embedding and masking strategies based on disease progression knowledge. Compared to popular baseline models and standard longitudinal Transformers, these simple yet effective extensions significantly enhance the predictive ability of deep classification models.
LMT: Longitudinal Mixing Training, a Framework to Predict Disease Progression from a Single Image
Oct 16, 2023Abstract:Longitudinal imaging is able to capture both static anatomical structures and dynamic changes in disease progression toward earlier and better patient-specific pathology management. However, conventional approaches rarely take advantage of longitudinal information for detection and prediction purposes, especially for Diabetic Retinopathy (DR). In the past years, Mix-up training and pretext tasks with longitudinal context have effectively enhanced DR classification results and captured disease progression. In the meantime, a novel type of neural network named Neural Ordinary Differential Equation (NODE) has been proposed for solving ordinary differential equations, with a neural network treated as a black box. By definition, NODE is well suited for solving time-related problems. In this paper, we propose to combine these three aspects to detect and predict DR progression. Our framework, Longitudinal Mixing Training (LMT), can be considered both as a regularizer and as a pretext task that encodes the disease progression in the latent space. Additionally, we evaluate the trained model weights on a downstream task with a longitudinal context using standard and longitudinal pretext tasks. We introduce a new way to train time-aware models using $t_{mix}$, a weighted average time between two consecutive examinations. We compare our approach to standard mixing training on DR classification using OPHDIAT a longitudinal retinal Color Fundus Photographs (CFP) dataset. We were able to predict whether an eye would develop a severe DR in the following visit using a single image, with an AUC of 0.798 compared to baseline results of 0.641. Our results indicate that our longitudinal pretext task can learn the progression of DR disease and that introducing $t_{mix}$ augmentation is beneficial for time-aware models.
Longitudinal Self-supervised Learning Using Neural Ordinary Differential Equation
Oct 16, 2023Abstract:Longitudinal analysis in medical imaging is crucial to investigate the progressive changes in anatomical structures or disease progression over time. In recent years, a novel class of algorithms has emerged with the goal of learning disease progression in a self-supervised manner, using either pairs of consecutive images or time series of images. By capturing temporal patterns without external labels or supervision, longitudinal self-supervised learning (LSSL) has become a promising avenue. To better understand this core method, we explore in this paper the LSSL algorithm under different scenarios. The original LSSL is embedded in an auto-encoder (AE) structure. However, conventional self-supervised strategies are usually implemented in a Siamese-like manner. Therefore, (as a first novelty) in this study, we explore the use of Siamese-like LSSL. Another new core framework named neural ordinary differential equation (NODE). NODE is a neural network architecture that learns the dynamics of ordinary differential equations (ODE) through the use of neural networks. Many temporal systems can be described by ODE, including modeling disease progression. We believe that there is an interesting connection to make between LSSL and NODE. This paper aims at providing a better understanding of those core algorithms for learning the disease progression with the mentioned change. In our different experiments, we employ a longitudinal dataset, named OPHDIAT, targeting diabetic retinopathy (DR) follow-up. Our results demonstrate the application of LSSL without including a reconstruction term, as well as the potential of incorporating NODE in conjunction with LSSL.
Detection of diabetic retinopathy using longitudinal self-supervised learning
Sep 05, 2022



Abstract:Longitudinal imaging is able to capture both static anatomical structures and dynamic changes in disease progression towards earlier and better patient-specific pathology management. However, conventional approaches for detecting diabetic retinopathy (DR) rarely take advantage of longitudinal information to improve DR analysis. In this work, we investigate the benefit of exploiting self-supervised learning with a longitudinal nature for DR diagnosis purposes. We compare different longitudinal self-supervised learning (LSSL) methods to model the disease progression from longitudinal retinal color fundus photographs (CFP) to detect early DR severity changes using a pair of consecutive exams. The experiments were conducted on a longitudinal DR screening dataset with or without those trained encoders (LSSL) acting as a longitudinal pretext task. Results achieve an AUC of 0.875 for the baseline (model trained from scratch) and an AUC of 0.96 (95% CI: 0.9593-0.9655 DeLong test) with a p-value < 2.2e-16 on early fusion using a simple ResNet alike architecture with frozen LSSL weights, suggesting that the LSSL latent space enables to encode the dynamic of DR progression.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge