Ahror Belaid
Arabic Handwritten Text Line Dataset
Dec 10, 2023Abstract:Segmentation of Arabic manuscripts into lines of text and words is an important step to make recognition systems more efficient and accurate. The problem of segmentation into text lines is solved since there are carefully annotated dataset dedicated to this task. However, To the best of our knowledge, there are no dataset annotating the word position of Arabic texts. In this paper, we present a new dataset specifically designed for historical Arabic script in which we annotate position in word level.
Cross-dimensional transfer learning in medical image segmentation with deep learning
Jul 29, 2023Abstract:Over the last decade, convolutional neural networks have emerged and advanced the state-of-the-art in various image analysis and computer vision applications. The performance of 2D image classification networks is constantly improving and being trained on databases made of millions of natural images. However, progress in medical image analysis has been hindered by limited annotated data and acquisition constraints. These limitations are even more pronounced given the volumetry of medical imaging data. In this paper, we introduce an efficient way to transfer the efficiency of a 2D classification network trained on natural images to 2D, 3D uni- and multi-modal medical image segmentation applications. In this direction, we designed novel architectures based on two key principles: weight transfer by embedding a 2D pre-trained encoder into a higher dimensional U-Net, and dimensional transfer by expanding a 2D segmentation network into a higher dimension one. The proposed networks were tested on benchmarks comprising different modalities: MR, CT, and ultrasound images. Our 2D network ranked first on the CAMUS challenge dedicated to echo-cardiographic data segmentation and surpassed the state-of-the-art. Regarding 2D/3D MR and CT abdominal images from the CHAOS challenge, our approach largely outperformed the other 2D-based methods described in the challenge paper on Dice, RAVD, ASSD, and MSSD scores and ranked third on the online evaluation platform. Our 3D network applied to the BraTS 2022 competition also achieved promising results, reaching an average Dice score of 91.69% (91.22%) for the whole tumor, 83.23% (84.77%) for the tumor core, and 81.75% (83.88%) for enhanced tumor using the approach based on weight (dimensional) transfer. Experimental and qualitative results illustrate the effectiveness of our methods for multi-dimensional medical image segmentation.
* 30 pages, 12 figures, 6 tables, Accepted for publication in the Journal of Medical Image Analysis
Efficient embedding network for 3D brain tumor segmentation
Nov 22, 2020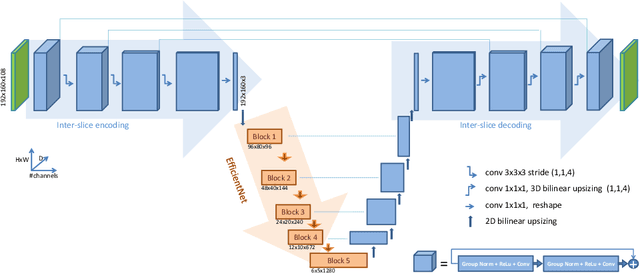
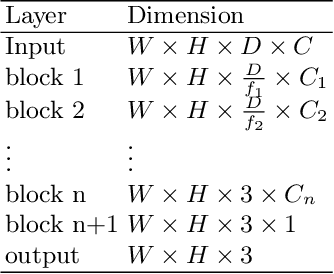
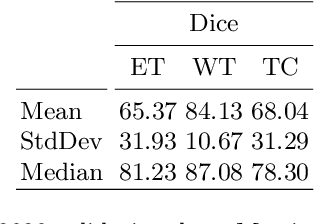
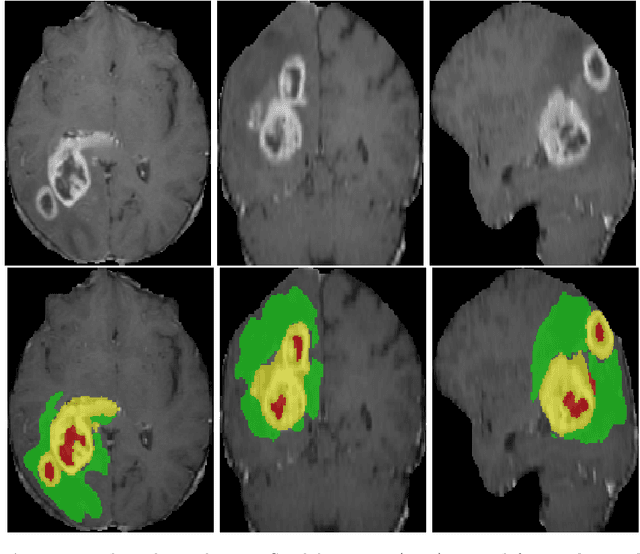
Abstract:3D medical image processing with deep learning greatly suffers from a lack of data. Thus, studies carried out in this field are limited compared to works related to 2D natural image analysis, where very large datasets exist. As a result, powerful and efficient 2D convolutional neural networks have been developed and trained. In this paper, we investigate a way to transfer the performance of a two-dimensional classiffication network for the purpose of three-dimensional semantic segmentation of brain tumors. We propose an asymmetric U-Net network by incorporating the EfficientNet model as part of the encoding branch. As the input data is in 3D, the first layers of the encoder are devoted to the reduction of the third dimension in order to fit the input of the EfficientNet network. Experimental results on validation and test data from the BraTS 2020 challenge demonstrate that the proposed method achieve promising performance.
* Multimodal Brain Tumor Segmentation Challenge 2020
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge