Odyssée Merveille
CREATIS
The R-Vessel-X Project
Jan 17, 2025



Abstract:1) Objectives: This technical report presents a synthetic summary and the principal outcomes of the project R-Vessel-X ("Robust vascular network extraction and understanding within hepatic biomedical images") funded by the French Agence Nationale de la Recherche, and developed between 2019 and 2023. 2) Material and methods: We used datasets and tools publicly available such as IRCAD, Bullitt or VascuSynth toobtain real or synthetic angiographic images. The main contributions lie in the field of 3D angiographic image analysis: filtering, segmentation, modeling and simulation, with a specific focus on the liver. 3) Results: We paid a particular attention to open-source software diffusion of the developed methods, by means of 3D Slicer plugins for the liver anatomy segmentation (SlicerRVXLiverSegmentation) and vesselness filtering (Slicer-RVXVesselnessFilters), and an online demo for the generation of synthetic and realistic vessels in 2D and 3D (OpenCCO). 4) Conclusion: The R-Vessel-X project provided extensive research outcomes, covering various topics related to 3D angiographic image analysis, such as filtering, segmentation, modeling and simulation. We also developed open-source and free softwares so that the research communities in biomedical engineering can use these results in their future research.
A plug-and-play framework for curvilinear structure segmentation based on a learned reconnecting regularization
Aug 23, 2024Abstract:Curvilinear structures are present in various fields in image processing such as blood vessels in medical imaging or roads in remote sensing. Their detection is crucial for many applications. In this article, we propose an unsupervised plug-and-play framework for the segmentation of curvilinear structures that focuses on the preservation of their connectivity. This framework includes an algorithm for generating realistic pairs of connected/disconnected curvilinear structures and a reconnecting regularization operator that can be learned from a synthetic dataset. Once learned, this regularization operator can be plugged into a variational segmentation scheme and used to segment curvilinear structure images without requiring annotations. We demonstrate the interest of our approach on the segmentation of vascular images both in 2D and 3D and compare its results with classic unsupervised and deep learning-based approach. Comparative evaluations against unsupervised classic and deep learning-based methods highlight the superior performance of our approach, showcasing remarkable improvements in preserving the connectivity of curvilinear structures (approximately 90% in 2D and 70% in 3D). We finally showcase the good generalizability behavior of our approach on two different applications : road cracks and porcine corneal cells segmentations.
Restoring Connectivity in Vascular Segmentation using a Learned Post-Processing Model
Apr 16, 2024Abstract:Accurate segmentation of vascular networks is essential for computer-aided tools designed to address cardiovascular diseases. Despite more than thirty years of research, it remains a challenge to obtain vascular segmentation results that preserve the connectivity of the underlying vascular network. Yet connectivity is one of the key feature of these tools. In this work, we propose a post-processing algorithm aiming to reconnect vascular structures that have been disconnected by a segmentation algorithm. Connectivity being a complex property to model explicity, we propose to learn this geometric feature either through synthetic data or annotations of the application of interest. The resulting post-processing model can be used on the output of any supervised or unsupervised vascular segmentation algorithm. We show that this post-processing effectively restores the connectivity of vascular networks both in 2D and 3D images, leading to improved overall segmentation results.
Guidelines for Cerebrovascular Segmentation: Managing Imperfect Annotations in the context of Semi-Supervised Learning
Apr 02, 2024Abstract:Segmentation in medical imaging is an essential and often preliminary task in the image processing chain, driving numerous efforts towards the design of robust segmentation algorithms. Supervised learning methods achieve excellent performances when fed with a sufficient amount of labeled data. However, such labels are typically highly time-consuming, error-prone and expensive to produce. Alternatively, semi-supervised learning approaches leverage both labeled and unlabeled data, and are very useful when only a small fraction of the dataset is labeled. They are particularly useful for cerebrovascular segmentation, given that labeling a single volume requires several hours for an expert. In addition to the challenge posed by insufficient annotations, there are concerns regarding annotation consistency. The task of annotating the cerebrovascular tree is inherently ambiguous. Due to the discrete nature of images, the borders and extremities of vessels are often unclear. Consequently, annotations heavily rely on the expert subjectivity and on the underlying clinical objective. These discrepancies significantly increase the complexity of the segmentation task for the model and consequently impair the results. Consequently, it becomes imperative to provide clinicians with precise guidelines to improve the annotation process and construct more uniform datasets. In this article, we investigate the data dependency of deep learning methods within the context of imperfect data and semi-supervised learning, for cerebrovascular segmentation. Specifically, this study compares various state-of-the-art semi-supervised methods based on unsupervised regularization and evaluates their performance in diverse quantity and quality data scenarios. Based on these experiments, we provide guidelines for the annotation and training of cerebrovascular segmentation models.
Cascaded multitask U-Net using topological loss for vessel segmentation and centerline extraction
Jul 21, 2023Abstract:Vessel segmentation and centerline extraction are two crucial preliminary tasks for many computer-aided diagnosis tools dealing with vascular diseases. Recently, deep-learning based methods have been widely applied to these tasks. However, classic deep-learning approaches struggle to capture the complex geometry and specific topology of vascular networks, which is of the utmost importance in most applications. To overcome these limitations, the clDice loss, a topological loss that focuses on the vessel centerlines, has been recently proposed. This loss requires computing, with a proposed soft-skeleton algorithm, the skeletons of both the ground truth and the predicted segmentation. However, the soft-skeleton algorithm provides suboptimal results on 3D images, which makes the clDice hardly suitable on 3D images. In this paper, we propose to replace the soft-skeleton algorithm by a U-Net which computes the vascular skeleton directly from the segmentation. We show that our method provides more accurate skeletons than the soft-skeleton algorithm. We then build upon this network a cascaded U-Net trained with the clDice loss to embed topological constraints during the segmentation. The resulting model is able to predict both the vessel segmentation and centerlines with a more accurate topology.
Strategies for Training Stain Invariant CNNs
Nov 03, 2018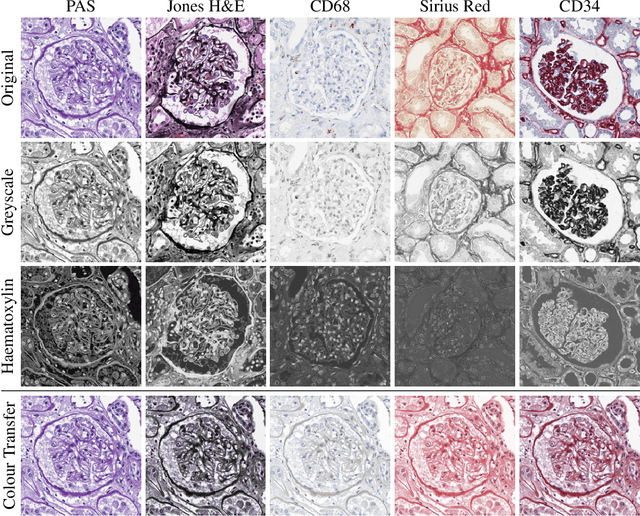
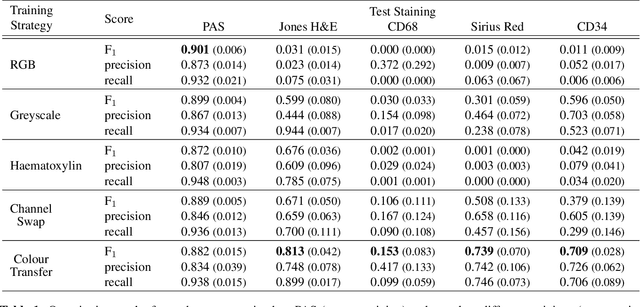
Abstract:An important part of Digital Pathology is the analysis of multiple digitised whole slide images from differently stained tissue sections. It is common practice to mount consecutive sections containing corresponding microscopic structures on glass slides, and to stain them differently to highlight specific tissue components. These multiple staining modalities result in very different images but include a significant amount of consistent image information. Deep learning approaches have recently been proposed to analyse these images in order to automatically identify objects of interest for pathologists. These supervised approaches require a vast amount of annotations, which are difficult and expensive to acquire---a problem that is multiplied with multiple stainings. This article presents several training strategies that make progress towards stain invariant networks. By training the network on one commonly used staining modality and applying it to images that include corresponding but differently stained tissue structures, the presented unsupervised strategies demonstrate significant improvements over standard training strategies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge