Jessica Schmitz
An automatic framework to study the tissue micro-environment of renal glomeruli in differently stained consecutive digital whole slide images
Aug 29, 2020



Abstract:Objective: This article presents an automatic image processing framework to extract quantitative high-level information describing the micro-environment of glomeruli in consecutive whole slide images (WSIs) processed with different staining modalities of patients with chronic kidney rejection after kidney transplantation. Methods: This three step framework consists of: 1) cell and anatomical structure segmentation based on colour deconvolution and deep learning 2) fusion of information from different stainings using a newly developed registration algorithm 3) feature extraction. Results: Each step of the framework is validated independently both quantitatively and qualitatively by pathologists. An illustration of the different types of features that can be extracted is presented. Conclusion: The proposed generic framework allows for the analysis of the micro-environment surrounding large structures that can be segmented (either manually or automatically). It is independent of the segmentation approach and is therefore applicable to a variety of biomedical research questions. Significance: Chronic tissue remodelling processes after kidney transplantation can result in interstitial fibrosis and tubular atrophy (IFTA) and glomerulosclerosis. This pipeline provides tools to quantitatively analyse, in the same spatial context, information from different consecutive WSIs and help researchers understand the complex underlying mechanisms leading to IFTA and glomerulosclerosis.
Strategies for Training Stain Invariant CNNs
Nov 03, 2018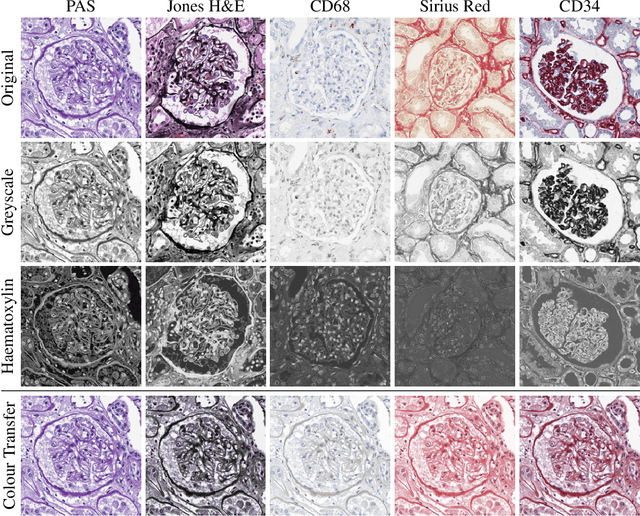
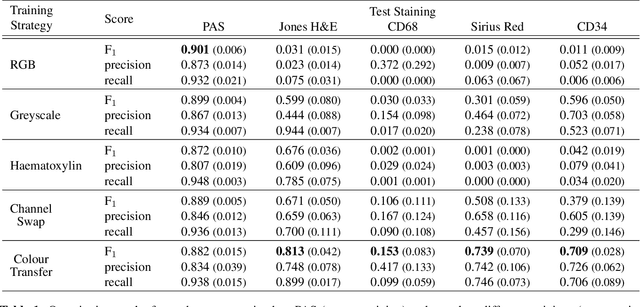
Abstract:An important part of Digital Pathology is the analysis of multiple digitised whole slide images from differently stained tissue sections. It is common practice to mount consecutive sections containing corresponding microscopic structures on glass slides, and to stain them differently to highlight specific tissue components. These multiple staining modalities result in very different images but include a significant amount of consistent image information. Deep learning approaches have recently been proposed to analyse these images in order to automatically identify objects of interest for pathologists. These supervised approaches require a vast amount of annotations, which are difficult and expensive to acquire---a problem that is multiplied with multiple stainings. This article presents several training strategies that make progress towards stain invariant networks. By training the network on one commonly used staining modality and applying it to images that include corresponding but differently stained tissue structures, the presented unsupervised strategies demonstrate significant improvements over standard training strategies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge