Dimitris Visvikis
LaTIM
Multi-Branch Generative Models for Multichannel Imaging with an Application to PET/CT Joint Reconstruction
Apr 12, 2024Abstract:This paper presents a proof-of-concept approach for learned synergistic reconstruction of medical images using multi-branch generative models. Leveraging variational autoencoders (VAEs) and generative adversarial networks (GANs), our models learn from pairs of images simultaneously, enabling effective denoising and reconstruction. Synergistic image reconstruction is achieved by incorporating the trained models in a regularizer that evaluates the distance between the images and the model, in a similar fashion to multichannel dictionary learning (DiL). We demonstrate the efficacy of our approach on both Modified National Institute of Standards and Technology (MNIST) and positron emission tomography (PET)/computed tomography (CT) datasets, showcasing improved image quality and information sharing between modalities. Despite challenges such as patch decomposition and model limitations, our results underscore the potential of generative models for enhancing medical imaging reconstruction.
CT respiratory motion synthesis using joint supervised and adversarial learning
Mar 29, 2024Abstract:Objective: Four-dimensional computed tomography (4DCT) imaging consists in reconstructing a CT acquisition into multiple phases to track internal organ and tumor motion. It is commonly used in radiotherapy treatment planning to establish planning target volumes. However, 4DCT increases protocol complexity, may not align with patient breathing during treatment, and lead to higher radiation delivery. Approach: In this study, we propose a deep synthesis method to generate pseudo respiratory CT phases from static images for motion-aware treatment planning. The model produces patient-specific deformation vector fields (DVFs) by conditioning synthesis on external patient surface-based estimation, mimicking respiratory monitoring devices. A key methodological contribution is to encourage DVF realism through supervised DVF training while using an adversarial term jointly not only on the warped image but also on the magnitude of the DVF itself. This way, we avoid excessive smoothness typically obtained through deep unsupervised learning, and encourage correlations with the respiratory amplitude. Main results: Performance is evaluated using real 4DCT acquisitions with smaller tumor volumes than previously reported. Results demonstrate for the first time that the generated pseudo-respiratory CT phases can capture organ and tumor motion with similar accuracy to repeated 4DCT scans of the same patient. Mean inter-scans tumor center-of-mass distances and Dice similarity coefficients were $1.97$mm and $0.63$, respectively, for real 4DCT phases and $2.35$mm and $0.71$ for synthetic phases, and compares favorably to a state-of-the-art technique (RMSim).
Cross-modal tumor segmentation using generative blending augmentation and self training
Apr 04, 2023Abstract:Deep learning for medical imaging is limited by data scarcity and domain shift, which lead to biased training sets that do not accurately represent deployment conditions. A related practical problem is cross-modal segmentation where the objective is to segment unlabelled domains using previously labelled images from other modalites, which is the context of the MICCAI CrossMoDA 2022 challenge on vestibular schwannoma (VS) segmentation. In this context, we propose a VS segmentation method that leverages conventional image-to-image translation and segmentation using iterative self training combined to a dedicated data augmentation technique called Generative Blending Augmentation (GBA). GBA is based on a one-shot 2D SinGAN generative model that allows to realistically diversify target tumor appearances in a downstream segmentation model, improving its generalization power at test time. Our solution ranked first on the VS segmentation task during the validation and test phase of the CrossModa 2022 challenge.
Multi-Channel Convolutional Analysis Operator Learning for Dual-Energy CT Reconstruction
Mar 10, 2022Abstract:Objective. Dual-energy computed tomography (DECT) has the potential to improve contrast, reduce artifacts and the ability to perform material decomposition in advanced imaging applications. The increased number or measurements results with a higher radiation dose and it is therefore essential to reduce either number of projections per energy or the source X-ray intensity, but this makes tomographic reconstruction more ill-posed. Approach. We developed the multi-channel convolutional analysis operator learning (MCAOL) method to exploit common spatial features within attenuation images at different energies and we propose an optimization method which jointly reconstructs the attenuation images at low and high energies with a mixed norm regularization on the sparse features obtained by pre-trained convolutional filters through the convolutional analysis operator learning (CAOL) algorithm. Main results. Extensive experiments with simulated and real computed tomography (CT) data were performed to validate the effectiveness of the proposed methods and we reported increased reconstruction accuracy compared to CAOL and iterative methods with single and joint total-variation (TV) regularization. Significance. Qualitative and quantitative results on sparse-views and low-dose DECT demonstrate that the proposed MCAOL method outperforms both CAOL applied on each energy independently and several existing state-of-the-art model-based iterative reconstruction (MBIR) techniques, thus paving the way for dose reduction.
* 23 pages, 11 figures, published in the Physics in Medicine & Biology journal
Overview of the HECKTOR Challenge at MICCAI 2021: Automatic Head and Neck Tumor Segmentation and Outcome Prediction in PET/CT Images
Jan 11, 2022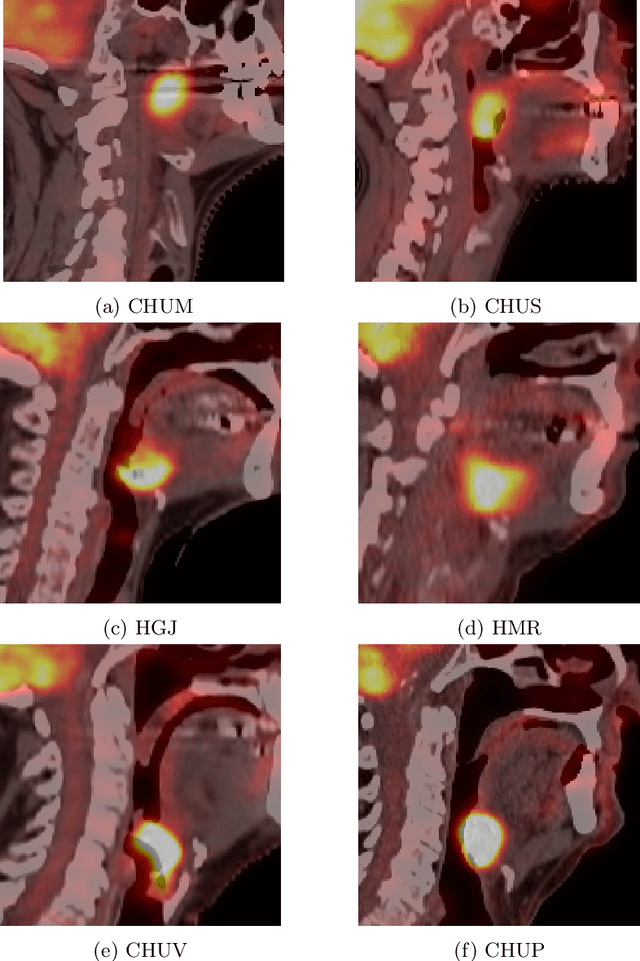
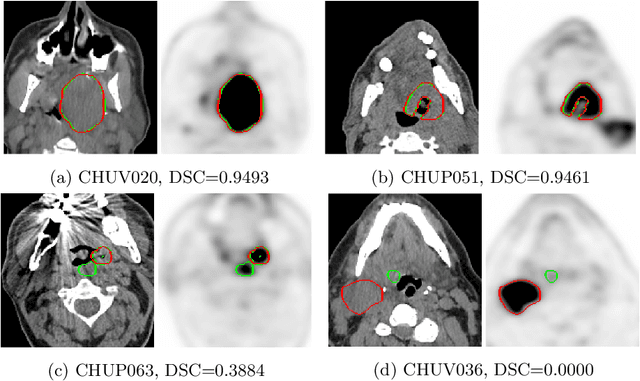
Abstract:This paper presents an overview of the second edition of the HEad and neCK TumOR (HECKTOR) challenge, organized as a satellite event of the 24th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) 2021. The challenge is composed of three tasks related to the automatic analysis of PET/CT images for patients with Head and Neck cancer (H&N), focusing on the oropharynx region. Task 1 is the automatic segmentation of H&N primary Gross Tumor Volume (GTVt) in FDG-PET/CT images. Task 2 is the automatic prediction of Progression Free Survival (PFS) from the same FDG-PET/CT. Finally, Task 3 is the same as Task 2 with ground truth GTVt annotations provided to the participants. The data were collected from six centers for a total of 325 images, split into 224 training and 101 testing cases. The interest in the challenge was highlighted by the important participation with 103 registered teams and 448 result submissions. The best methods obtained a Dice Similarity Coefficient (DSC) of 0.7591 in the first task, and a Concordance index (C-index) of 0.7196 and 0.6978 in Tasks 2 and 3, respectively. In all tasks, simplicity of the approach was found to be key to ensure generalization performance. The comparison of the PFS prediction performance in Tasks 2 and 3 suggests that providing the GTVt contour was not crucial to achieve best results, which indicates that fully automatic methods can be used. This potentially obviates the need for GTVt contouring, opening avenues for reproducible and large scale radiomics studies including thousands potential subjects.
Learn2Reg: comprehensive multi-task medical image registration challenge, dataset and evaluation in the era of deep learning
Dec 23, 2021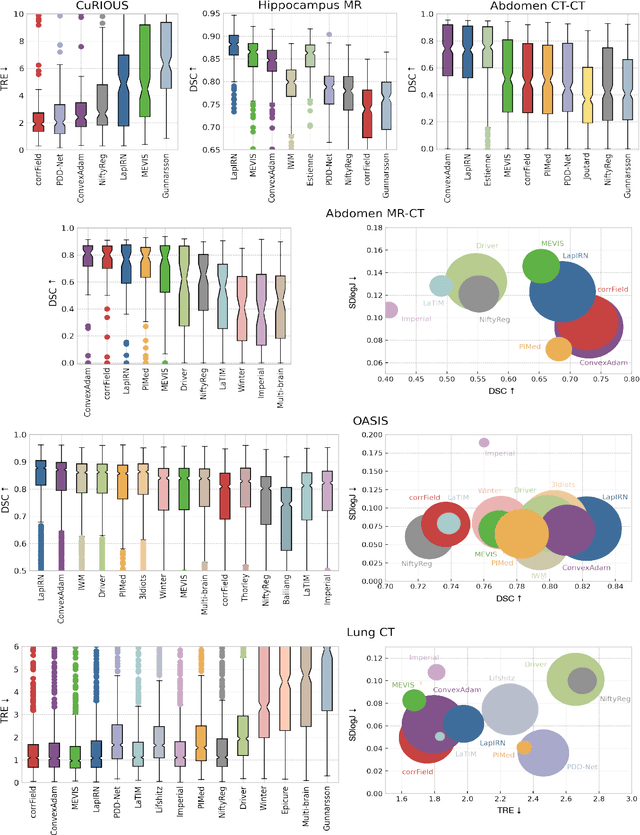
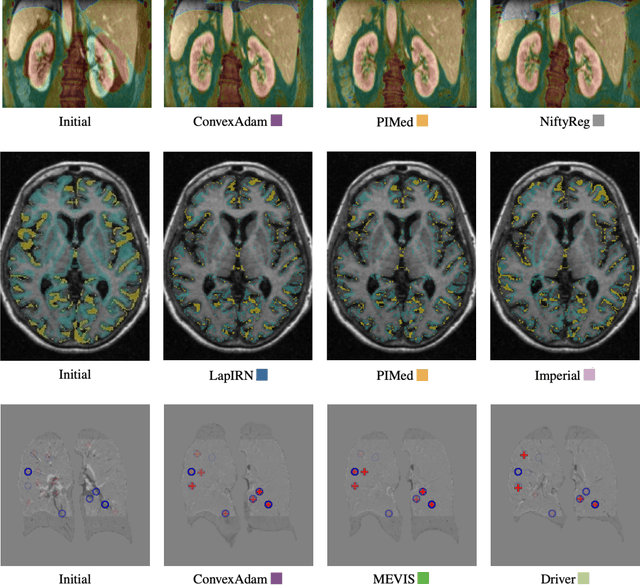
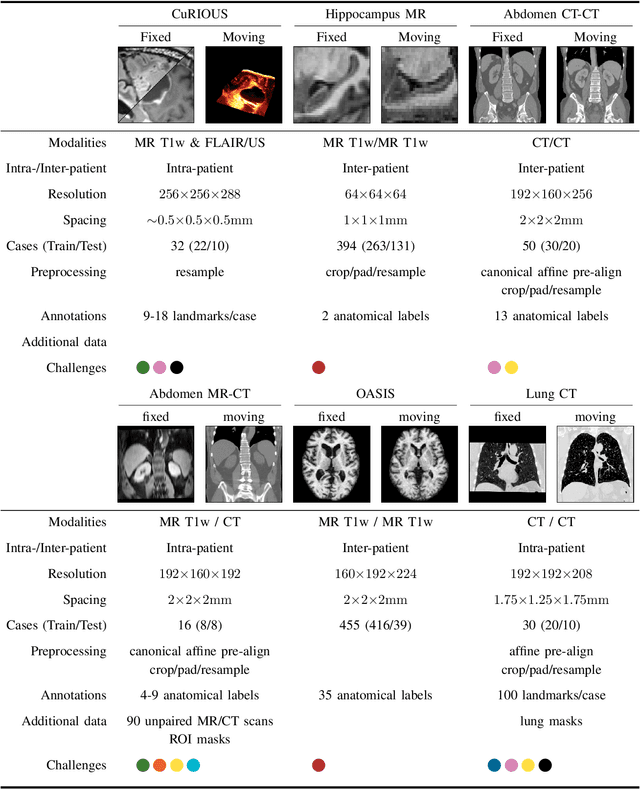
Abstract:Image registration is a fundamental medical image analysis task, and a wide variety of approaches have been proposed. However, only a few studies have comprehensively compared medical image registration approaches on a wide range of clinically relevant tasks, in part because of the lack of availability of such diverse data. This limits the development of registration methods, the adoption of research advances into practice, and a fair benchmark across competing approaches. The Learn2Reg challenge addresses these limitations by providing a multi-task medical image registration benchmark for comprehensive characterisation of deformable registration algorithms. A continuous evaluation will be possible at https://learn2reg.grand-challenge.org. Learn2Reg covers a wide range of anatomies (brain, abdomen, and thorax), modalities (ultrasound, CT, MR), availability of annotations, as well as intra- and inter-patient registration evaluation. We established an easily accessible framework for training and validation of 3D registration methods, which enabled the compilation of results of over 65 individual method submissions from more than 20 unique teams. We used a complementary set of metrics, including robustness, accuracy, plausibility, and runtime, enabling unique insight into the current state-of-the-art of medical image registration. This paper describes datasets, tasks, evaluation methods and results of the challenge, and the results of further analysis of transferability to new datasets, the importance of label supervision, and resulting bias.
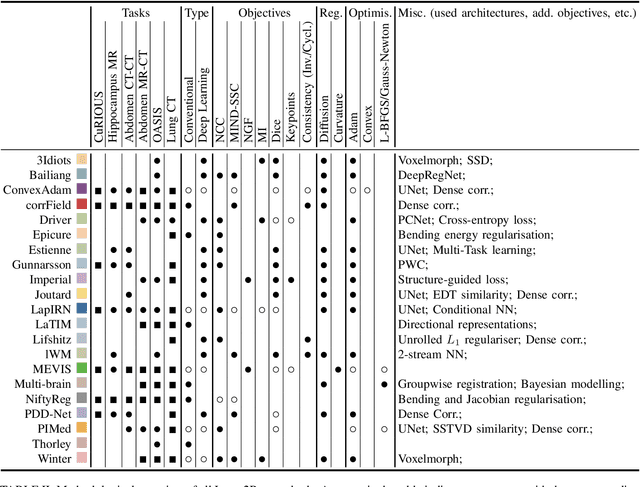
Regularized directional representations for medical image registration
Nov 30, 2021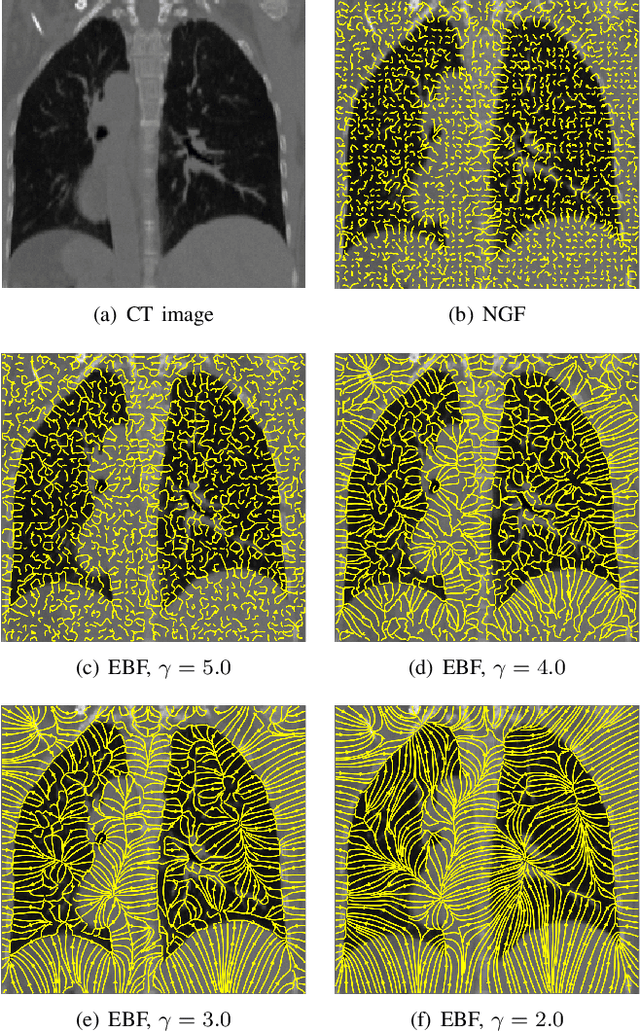
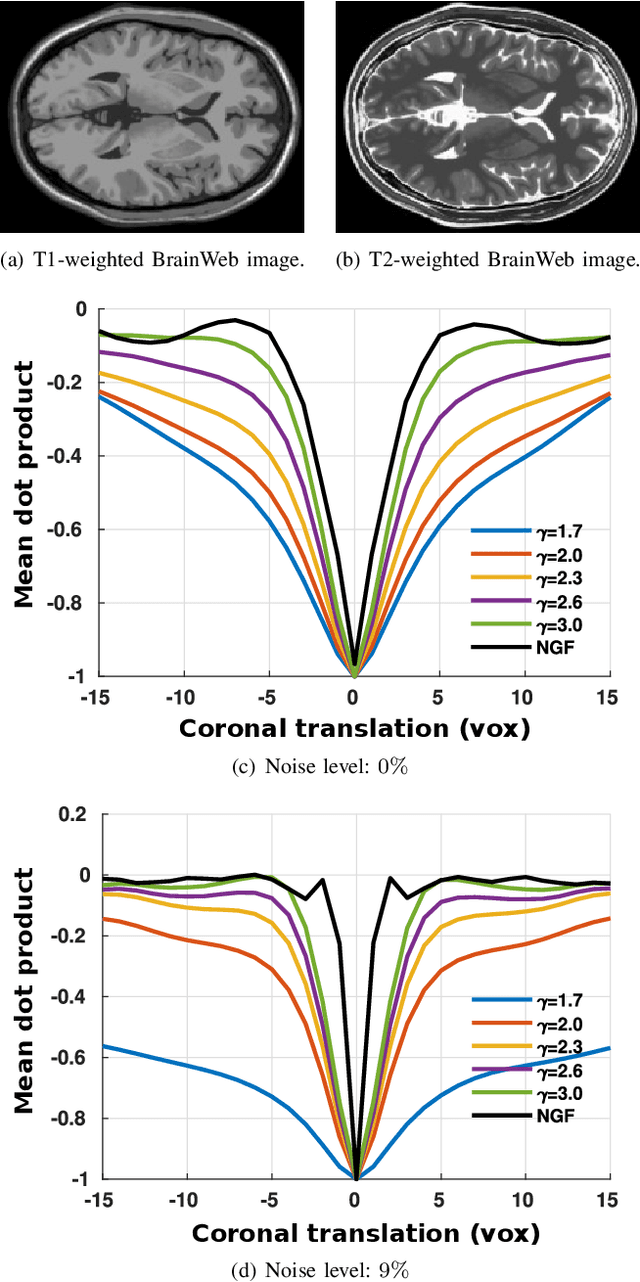
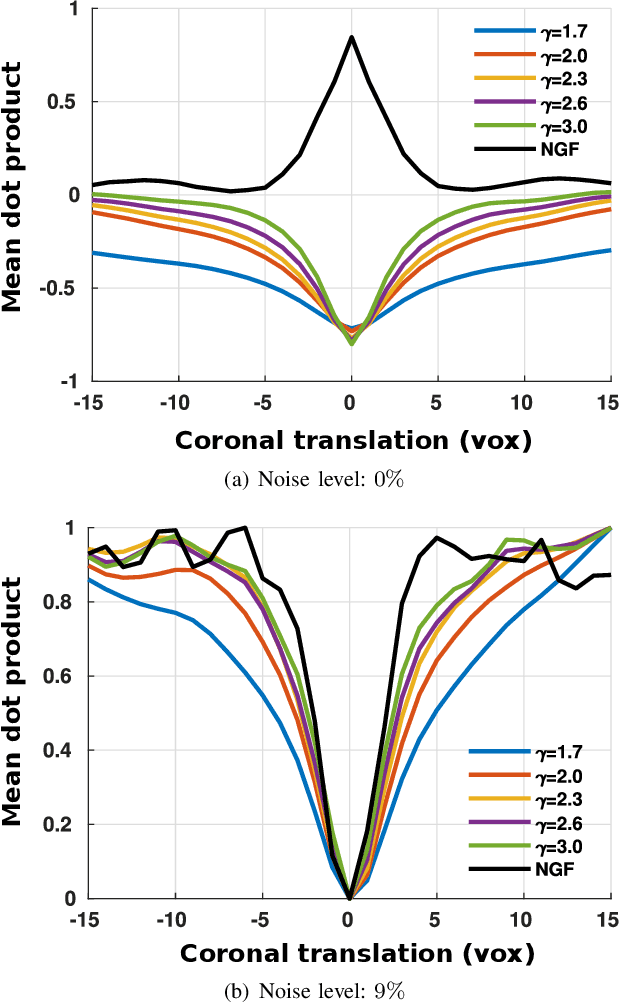
Abstract:In image registration, many efforts have been devoted to the development of alternatives to the popular normalized mutual information criterion. Concurrently to these efforts, an increasing number of works have demonstrated that substantial gains in registration accuracy can also be achieved by aligning structural representations of images rather than images themselves. Following this research path, we propose a new method for mono- and multimodal image registration based on the alignment of regularized vector fields derived from structural information such as gradient vector flow fields, a technique we call \textit{vector field similarity}. Our approach can be combined in a straightforward fashion with any existing registration framework by substituting vector field similarity to intensity-based registration. In our experiments, we show that the proposed approach compares favourably with conventional image alignment on several public image datasets using a diversity of imaging modalities and anatomical locations.
Squeeze-and-Excitation Normalization for Automated Delineation of Head and Neck Primary Tumors in Combined PET and CT Images
Feb 20, 2021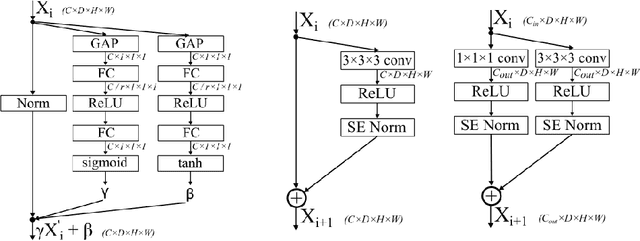
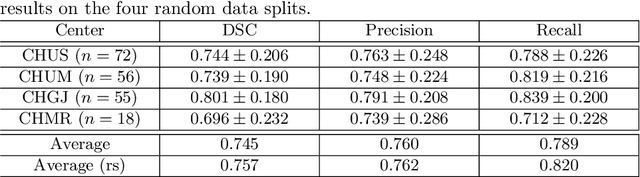
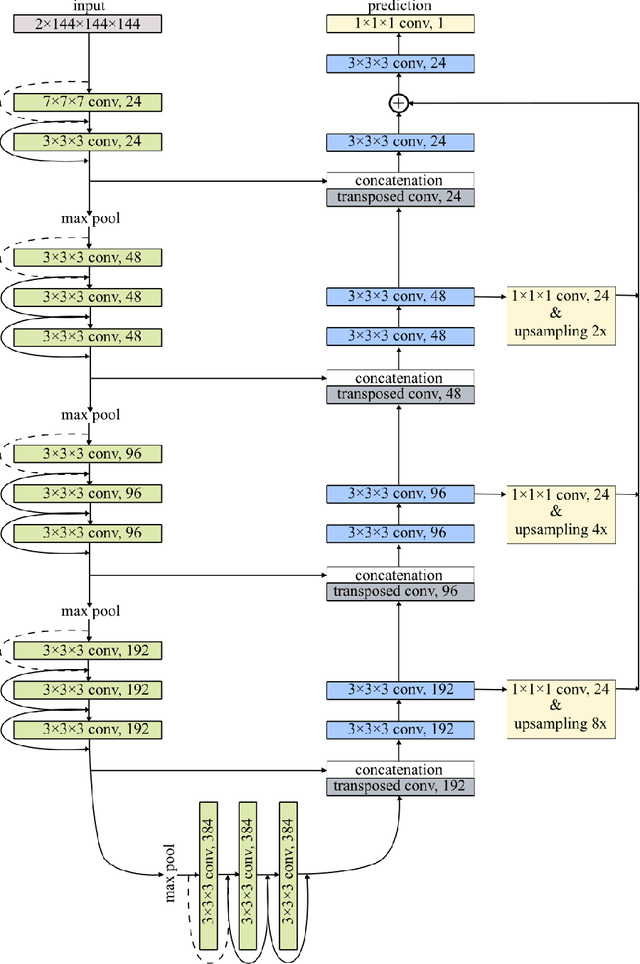

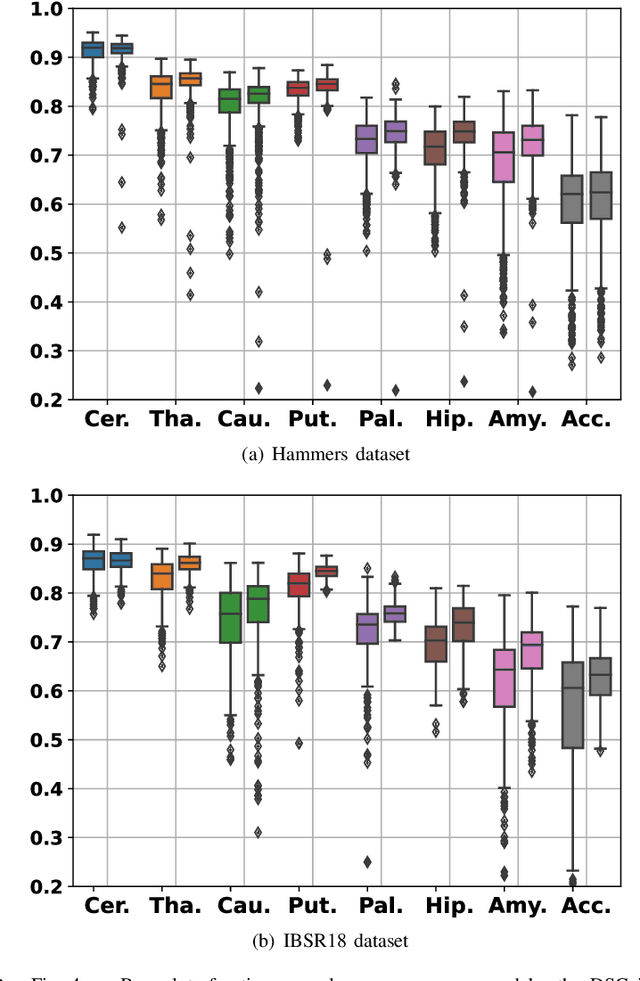
Abstract:Development of robust and accurate fully automated methods for medical image segmentation is crucial in clinical practice and radiomics studies. In this work, we contributed an automated approach for Head and Neck (H&N) primary tumor segmentation in combined positron emission tomography / computed tomography (PET/CT) images in the context of the MICCAI 2020 Head and Neck Tumor segmentation challenge (HECKTOR). Our model was designed on the U-Net architecture with residual layers and supplemented with Squeeze-and-Excitation Normalization. The described method achieved competitive results in cross-validation (DSC 0.745, precision 0.760, recall 0.789) performed on different centers, as well as on the test set (DSC 0.759, precision 0.833, recall 0.740) that allowed us to win first prize in the HECKTOR challenge among 21 participating teams. The full implementation based on PyTorch and the trained models are available at https://github.com/iantsen/hecktor
* 7 pages, 2 figures, 2 tables
Reliability of PET/CT shape and heterogeneity features in functional and morphological components of Non-Small Cell Lung Cancer tumors: a repeatability analysis in a prospective multi-center cohort
Oct 05, 2016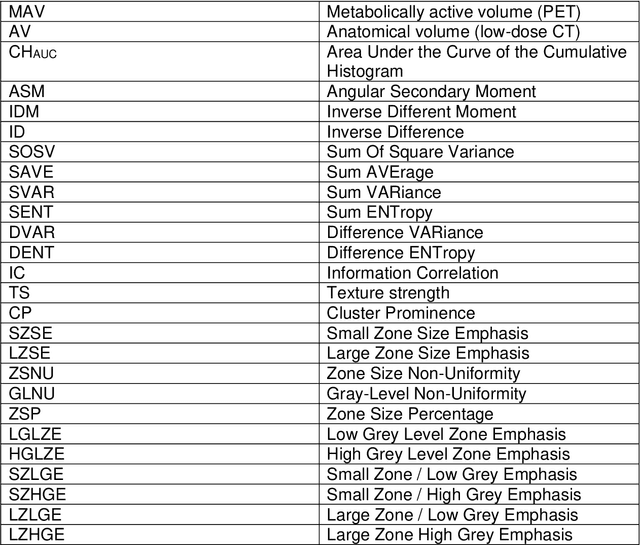
Abstract:Purpose: The main purpose of this study was to assess the reliability of shape and heterogeneity features in both Positron Emission Tomography (PET) and low-dose Computed Tomography (CT) components of PET/CT. A secondary objective was to investigate the impact of image quantization.Material and methods: A Health Insurance Portability and Accountability Act -compliant secondary analysis of deidentified prospectively acquired PET/CT test-retest datasets of 74 patients from multi-center Merck and ACRIN trials was performed. Metabolically active volumes were automatically delineated on PET with Fuzzy Locally Adaptive Bayesian algorithm. 3DSlicerTM was used to semi-automatically delineate the anatomical volumes on low-dose CT components. Two quantization methods were considered: a quantization into a set number of bins (quantizationB) and an alternative quantization with bins of fixed width (quantizationW). Four shape descriptors, ten first-order metrics and 26 textural features were computed. Bland-Altman analysis was used to quantify repeatability. Features were subsequently categorized as very reliable, reliable, moderately reliable and poorly reliable with respect to the corresponding volume variability. Results: Repeatability was highly variable amongst features. Numerous metrics were identified as poorly or moderately reliable. Others were (very) reliable in both modalities, and in all categories (shape, 1st-, 2nd- and 3rd-order metrics). Image quantization played a major role in the features repeatability. Features were more reliable in PET with quantizationB, whereas quantizationW showed better results in CT.Conclusion: The test-retest repeatability of shape and heterogeneity features in PET and low-dose CT varied greatly amongst metrics. The level of repeatability also depended strongly on the quantization step, with different optimal choices for each modality. The repeatability of PET and low-dose CT features should be carefully taken into account when selecting metrics to build multiparametric models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge