Siyuan Yan
A Vision-Language Foundation Model for Zero-shot Clinical Collaboration and Automated Concept Discovery in Dermatology
Feb 11, 2026Abstract:Medical foundation models have shown promise in controlled benchmarks, yet widespread deployment remains hindered by reliance on task-specific fine-tuning. Here, we introduce DermFM-Zero, a dermatology vision-language foundation model trained via masked latent modelling and contrastive learning on over 4 million multimodal data points. We evaluated DermFM-Zero across 20 benchmarks spanning zero-shot diagnosis and multimodal retrieval, achieving state-of-the-art performance without task-specific adaptation. We further evaluated its zero-shot capabilities in three multinational reader studies involving over 1,100 clinicians. In primary care settings, AI assistance enabled general practitioners to nearly double their differential diagnostic accuracy across 98 skin conditions. In specialist settings, the model significantly outperformed board-certified dermatologists in multimodal skin cancer assessment. In collaborative workflows, AI assistance enabled non-experts to surpass unassisted experts while improving management appropriateness. Finally, we show that DermFM-Zero's latent representations are interpretable: sparse autoencoders unsupervisedly disentangle clinically meaningful concepts that outperform predefined-vocabulary approaches and enable targeted suppression of artifact-induced biases, enhancing robustness without retraining. These findings demonstrate that a foundation model can provide effective, safe, and transparent zero-shot clinical decision support.
DermoGPT: Open Weights and Open Data for Morphology-Grounded Dermatological Reasoning MLLMs
Jan 05, 2026Abstract:Multimodal Large Language Models (MLLMs) show promise for medical applications, yet progress in dermatology lags due to limited training data, narrow task coverage, and lack of clinically-grounded supervision that mirrors expert diagnostic workflows. We present a comprehensive framework to address these gaps. First, we introduce DermoInstruct, a large-scale morphology-anchored instruction corpus comprising 211,243 images and 772,675 trajectories across five task formats, capturing the complete diagnostic pipeline from morphological observation and clinical reasoning to final diagnosis. Second, we establish DermoBench, a rigorous benchmark evaluating 11 tasks across four clinical axes: Morphology, Diagnosis, Reasoning, and Fairness, including a challenging subset of 3,600 expert-verified open-ended instances and human performance baselines. Third, we develop DermoGPT, a dermatology reasoning MLLM trained via supervised fine-tuning followed by our Morphologically-Anchored Visual-Inference-Consistent (MAVIC) reinforcement learning objective, which enforces consistency between visual observations and diagnostic conclusions. At inference, we deploy Confidence-Consistency Test-time adaptation (CCT) for robust predictions. Experiments show DermoGPT significantly outperforms 16 representative baselines across all axes, achieving state-of-the-art performance while substantially narrowing the human-AI gap. DermoInstruct, DermoBench and DermoGPT will be made publicly available at https://github.com/mendicant04/DermoGPT upon acceptance.
A Survey of Scientific Large Language Models: From Data Foundations to Agent Frontiers
Aug 28, 2025



Abstract:Scientific Large Language Models (Sci-LLMs) are transforming how knowledge is represented, integrated, and applied in scientific research, yet their progress is shaped by the complex nature of scientific data. This survey presents a comprehensive, data-centric synthesis that reframes the development of Sci-LLMs as a co-evolution between models and their underlying data substrate. We formulate a unified taxonomy of scientific data and a hierarchical model of scientific knowledge, emphasizing the multimodal, cross-scale, and domain-specific challenges that differentiate scientific corpora from general natural language processing datasets. We systematically review recent Sci-LLMs, from general-purpose foundations to specialized models across diverse scientific disciplines, alongside an extensive analysis of over 270 pre-/post-training datasets, showing why Sci-LLMs pose distinct demands -- heterogeneous, multi-scale, uncertainty-laden corpora that require representations preserving domain invariance and enabling cross-modal reasoning. On evaluation, we examine over 190 benchmark datasets and trace a shift from static exams toward process- and discovery-oriented assessments with advanced evaluation protocols. These data-centric analyses highlight persistent issues in scientific data development and discuss emerging solutions involving semi-automated annotation pipelines and expert validation. Finally, we outline a paradigm shift toward closed-loop systems where autonomous agents based on Sci-LLMs actively experiment, validate, and contribute to a living, evolving knowledge base. Collectively, this work provides a roadmap for building trustworthy, continually evolving artificial intelligence (AI) systems that function as a true partner in accelerating scientific discovery.
Controllable Skin Synthesis via Lesion-Focused Vector Autoregression Model
Aug 27, 2025Abstract:Skin images from real-world clinical practice are often limited, resulting in a shortage of training data for deep-learning models. While many studies have explored skin image synthesis, existing methods often generate low-quality images and lack control over the lesion's location and type. To address these limitations, we present LF-VAR, a model leveraging quantified lesion measurement scores and lesion type labels to guide the clinically relevant and controllable synthesis of skin images. It enables controlled skin synthesis with specific lesion characteristics based on language prompts. We train a multiscale lesion-focused Vector Quantised Variational Auto-Encoder (VQVAE) to encode images into discrete latent representations for structured tokenization. Then, a Visual AutoRegressive (VAR) Transformer trained on tokenized representations facilitates image synthesis. Lesion measurement from the lesion region and types as conditional embeddings are integrated to enhance synthesis fidelity. Our method achieves the best overall FID score (average 0.74) among seven lesion types, improving upon the previous state-of-the-art (SOTA) by 6.3%. The study highlights our controllable skin synthesis model's effectiveness in generating high-fidelity, clinically relevant synthetic skin images. Our framework code is available at https://github.com/echosun1996/LF-VAR.
MAKE: Multi-Aspect Knowledge-Enhanced Vision-Language Pretraining for Zero-shot Dermatological Assessment
May 14, 2025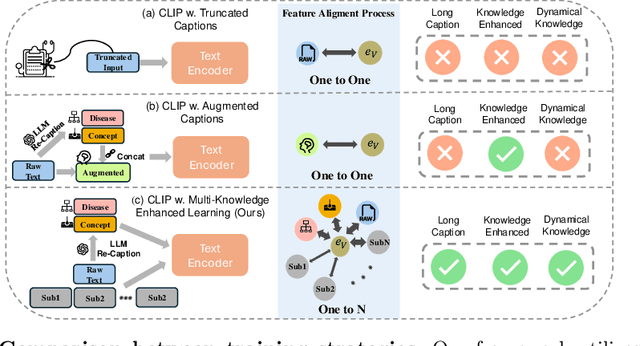
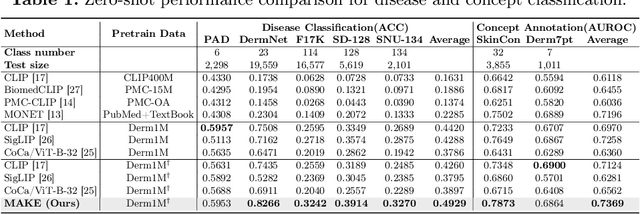
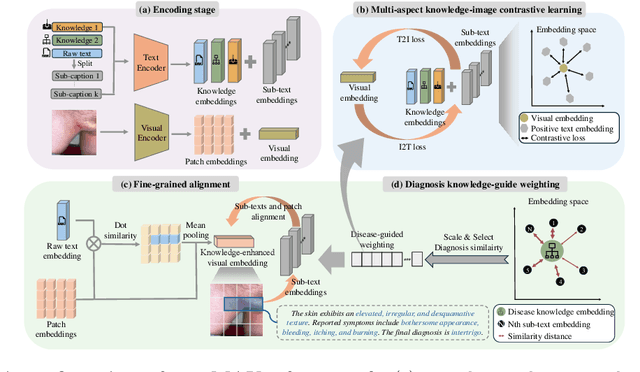
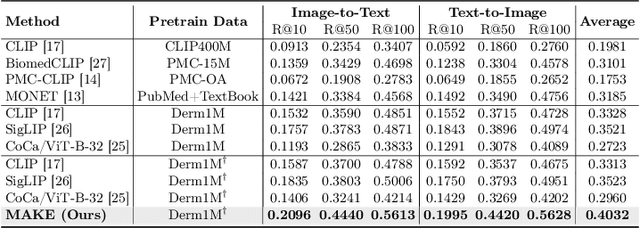
Abstract:Dermatological diagnosis represents a complex multimodal challenge that requires integrating visual features with specialized clinical knowledge. While vision-language pretraining (VLP) has advanced medical AI, its effectiveness in dermatology is limited by text length constraints and the lack of structured texts. In this paper, we introduce MAKE, a Multi-Aspect Knowledge-Enhanced vision-language pretraining framework for zero-shot dermatological tasks. Recognizing that comprehensive dermatological descriptions require multiple knowledge aspects that exceed standard text constraints, our framework introduces: (1) a multi-aspect contrastive learning strategy that decomposes clinical narratives into knowledge-enhanced sub-texts through large language models, (2) a fine-grained alignment mechanism that connects subcaptions with diagnostically relevant image features, and (3) a diagnosis-guided weighting scheme that adaptively prioritizes different sub-captions based on clinical significance prior. Through pretraining on 403,563 dermatological image-text pairs collected from education resources, MAKE significantly outperforms state-of-the-art VLP models on eight datasets across zero-shot skin disease classification, concept annotation, and cross-modal retrieval tasks. Our code will be made publicly available at https: //github.com/SiyuanYan1/MAKE.
Derm1M: A Million-scale Vision-Language Dataset Aligned with Clinical Ontology Knowledge for Dermatology
Mar 19, 2025Abstract:The emergence of vision-language models has transformed medical AI, enabling unprecedented advances in diagnostic capability and clinical applications. However, progress in dermatology has lagged behind other medical domains due to the lack of standard image-text pairs. Existing dermatological datasets are limited in both scale and depth, offering only single-label annotations across a narrow range of diseases instead of rich textual descriptions, and lacking the crucial clinical context needed for real-world applications. To address these limitations, we present Derm1M, the first large-scale vision-language dataset for dermatology, comprising 1,029,761 image-text pairs. Built from diverse educational resources and structured around a standard ontology collaboratively developed by experts, Derm1M provides comprehensive coverage for over 390 skin conditions across four hierarchical levels and 130 clinical concepts with rich contextual information such as medical history, symptoms, and skin tone. To demonstrate Derm1M potential in advancing both AI research and clinical application, we pretrained a series of CLIP-like models, collectively called DermLIP, on this dataset. The DermLIP family significantly outperforms state-of-the-art foundation models on eight diverse datasets across multiple tasks, including zero-shot skin disease classification, clinical and artifacts concept identification, few-shot/full-shot learning, and cross-modal retrieval. Our dataset and code will be public.
OphCLIP: Hierarchical Retrieval-Augmented Learning for Ophthalmic Surgical Video-Language Pretraining
Nov 23, 2024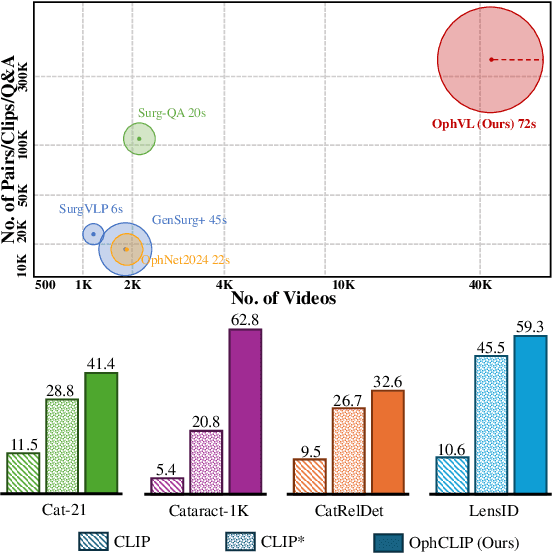
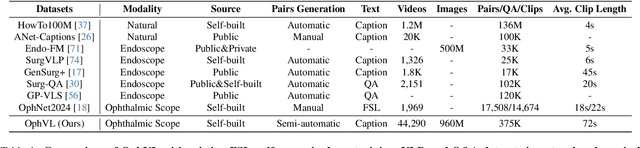
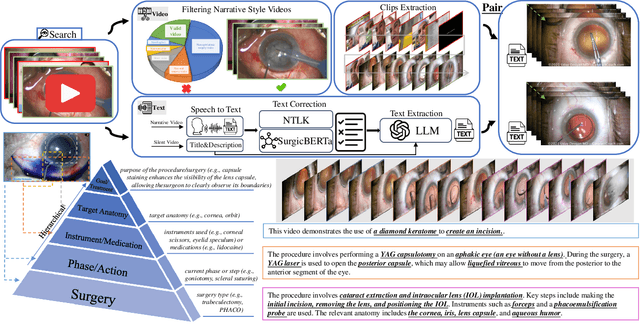
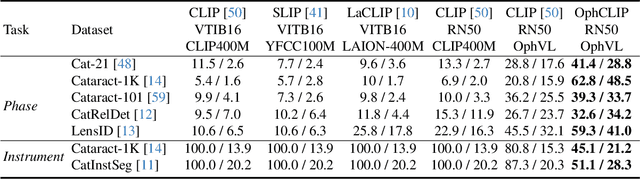
Abstract:Surgical practice involves complex visual interpretation, procedural skills, and advanced medical knowledge, making surgical vision-language pretraining (VLP) particularly challenging due to this complexity and the limited availability of annotated data. To address the gap, we propose OphCLIP, a hierarchical retrieval-augmented vision-language pretraining framework specifically designed for ophthalmic surgical workflow understanding. OphCLIP leverages the OphVL dataset we constructed, a large-scale and comprehensive collection of over 375K hierarchically structured video-text pairs with tens of thousands of different combinations of attributes (surgeries, phases/operations/actions, instruments, medications, as well as more advanced aspects like the causes of eye diseases, surgical objectives, and postoperative recovery recommendations, etc). These hierarchical video-text correspondences enable OphCLIP to learn both fine-grained and long-term visual representations by aligning short video clips with detailed narrative descriptions and full videos with structured titles, capturing intricate surgical details and high-level procedural insights, respectively. Our OphCLIP also designs a retrieval-augmented pretraining framework to leverage the underexplored large-scale silent surgical procedure videos, automatically retrieving semantically relevant content to enhance the representation learning of narrative videos. Evaluation across 11 datasets for phase recognition and multi-instrument identification shows OphCLIP's robust generalization and superior performance.
A General-Purpose Multimodal Foundation Model for Dermatology
Oct 19, 2024



Abstract:Diagnosing and treating skin diseases require advanced visual skills across multiple domains and the ability to synthesize information from various imaging modalities. Current deep learning models, while effective at specific tasks such as diagnosing skin cancer from dermoscopic images, fall short in addressing the complex, multimodal demands of clinical practice. Here, we introduce PanDerm, a multimodal dermatology foundation model pretrained through self-supervised learning on a dataset of over 2 million real-world images of skin diseases, sourced from 11 clinical institutions across 4 imaging modalities. We evaluated PanDerm on 28 diverse datasets covering a range of clinical tasks, including skin cancer screening, phenotype assessment and risk stratification, diagnosis of neoplastic and inflammatory skin diseases, skin lesion segmentation, change monitoring, and metastasis prediction and prognosis. PanDerm achieved state-of-the-art performance across all evaluated tasks, often outperforming existing models even when using only 5-10% of labeled data. PanDerm's clinical utility was demonstrated through reader studies in real-world clinical settings across multiple imaging modalities. It outperformed clinicians by 10.2% in early-stage melanoma detection accuracy and enhanced clinicians' multiclass skin cancer diagnostic accuracy by 11% in a collaborative human-AI setting. Additionally, PanDerm demonstrated robust performance across diverse demographic factors, including different body locations, age groups, genders, and skin tones. The strong results in benchmark evaluations and real-world clinical scenarios suggest that PanDerm could enhance the management of skin diseases and serve as a model for developing multimodal foundation models in other medical specialties, potentially accelerating the integration of AI support in healthcare.
MONICA: Benchmarking on Long-tailed Medical Image Classification
Oct 02, 2024



Abstract:Long-tailed learning is considered to be an extremely challenging problem in data imbalance learning. It aims to train well-generalized models from a large number of images that follow a long-tailed class distribution. In the medical field, many diagnostic imaging exams such as dermoscopy and chest radiography yield a long-tailed distribution of complex clinical findings. Recently, long-tailed learning in medical image analysis has garnered significant attention. However, the field currently lacks a unified, strictly formulated, and comprehensive benchmark, which often leads to unfair comparisons and inconclusive results. To help the community improve the evaluation and advance, we build a unified, well-structured codebase called Medical OpeN-source Long-taIled ClassifiCAtion (MONICA), which implements over 30 methods developed in relevant fields and evaluated on 12 long-tailed medical datasets covering 6 medical domains. Our work provides valuable practical guidance and insights for the field, offering detailed analysis and discussion on the effectiveness of individual components within the inbuilt state-of-the-art methodologies. We hope this codebase serves as a comprehensive and reproducible benchmark, encouraging further advancements in long-tailed medical image learning. The codebase is publicly available on https://github.com/PyJulie/MONICA.
OphNet: A Large-Scale Video Benchmark for Ophthalmic Surgical Workflow Understanding
Jun 12, 2024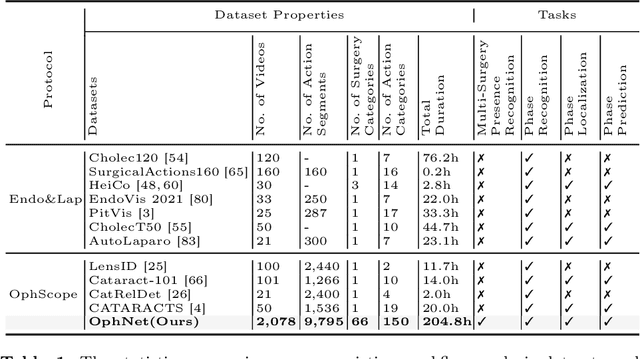
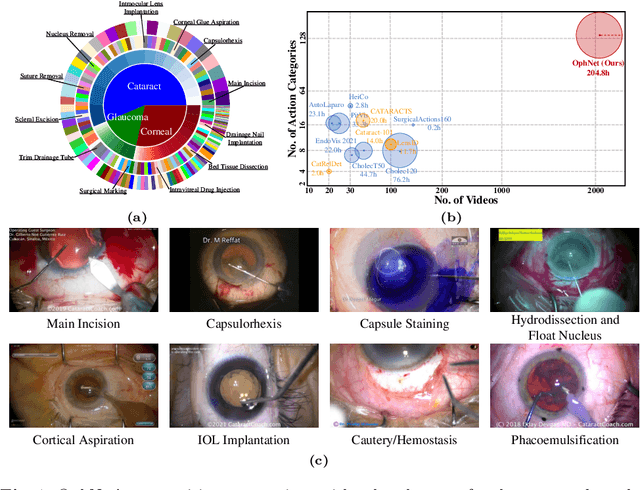

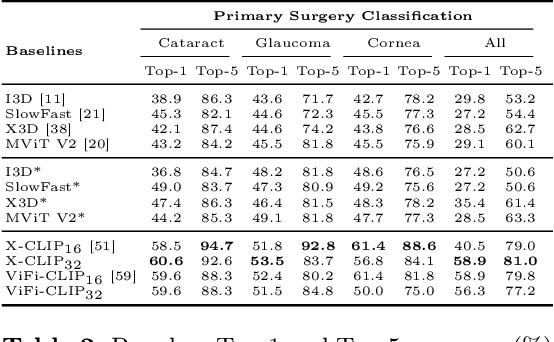
Abstract:Surgical scene perception via videos are critical for advancing robotic surgery, telesurgery, and AI-assisted surgery, particularly in ophthalmology. However, the scarcity of diverse and richly annotated video datasets has hindered the development of intelligent systems for surgical workflow analysis. Existing datasets for surgical workflow analysis, which typically face challenges such as small scale, a lack of diversity in surgery and phase categories, and the absence of time-localized annotations, limit the requirements for action understanding and model generalization validation in complex and diverse real-world surgical scenarios. To address this gap, we introduce OphNet, a large-scale, expert-annotated video benchmark for ophthalmic surgical workflow understanding. OphNet features: 1) A diverse collection of 2,278 surgical videos spanning 66 types of cataract, glaucoma, and corneal surgeries, with detailed annotations for 102 unique surgical phases and 150 granular operations; 2) It offers sequential and hierarchical annotations for each surgery, phase, and operation, enabling comprehensive understanding and improved interpretability; 3) Moreover, OphNet provides time-localized annotations, facilitating temporal localization and prediction tasks within surgical workflows. With approximately 205 hours of surgical videos, OphNet is about 20 times larger than the largest existing surgical workflow analysis benchmark. Our dataset and code have been made available at: \url{https://github.com/minghu0830/OphNet-benchmark}.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge