Harald Kittler
Derm1M: A Million-scale Vision-Language Dataset Aligned with Clinical Ontology Knowledge for Dermatology
Mar 19, 2025Abstract:The emergence of vision-language models has transformed medical AI, enabling unprecedented advances in diagnostic capability and clinical applications. However, progress in dermatology has lagged behind other medical domains due to the lack of standard image-text pairs. Existing dermatological datasets are limited in both scale and depth, offering only single-label annotations across a narrow range of diseases instead of rich textual descriptions, and lacking the crucial clinical context needed for real-world applications. To address these limitations, we present Derm1M, the first large-scale vision-language dataset for dermatology, comprising 1,029,761 image-text pairs. Built from diverse educational resources and structured around a standard ontology collaboratively developed by experts, Derm1M provides comprehensive coverage for over 390 skin conditions across four hierarchical levels and 130 clinical concepts with rich contextual information such as medical history, symptoms, and skin tone. To demonstrate Derm1M potential in advancing both AI research and clinical application, we pretrained a series of CLIP-like models, collectively called DermLIP, on this dataset. The DermLIP family significantly outperforms state-of-the-art foundation models on eight diverse datasets across multiple tasks, including zero-shot skin disease classification, clinical and artifacts concept identification, few-shot/full-shot learning, and cross-modal retrieval. Our dataset and code will be public.
A General-Purpose Multimodal Foundation Model for Dermatology
Oct 19, 2024



Abstract:Diagnosing and treating skin diseases require advanced visual skills across multiple domains and the ability to synthesize information from various imaging modalities. Current deep learning models, while effective at specific tasks such as diagnosing skin cancer from dermoscopic images, fall short in addressing the complex, multimodal demands of clinical practice. Here, we introduce PanDerm, a multimodal dermatology foundation model pretrained through self-supervised learning on a dataset of over 2 million real-world images of skin diseases, sourced from 11 clinical institutions across 4 imaging modalities. We evaluated PanDerm on 28 diverse datasets covering a range of clinical tasks, including skin cancer screening, phenotype assessment and risk stratification, diagnosis of neoplastic and inflammatory skin diseases, skin lesion segmentation, change monitoring, and metastasis prediction and prognosis. PanDerm achieved state-of-the-art performance across all evaluated tasks, often outperforming existing models even when using only 5-10% of labeled data. PanDerm's clinical utility was demonstrated through reader studies in real-world clinical settings across multiple imaging modalities. It outperformed clinicians by 10.2% in early-stage melanoma detection accuracy and enhanced clinicians' multiclass skin cancer diagnostic accuracy by 11% in a collaborative human-AI setting. Additionally, PanDerm demonstrated robust performance across diverse demographic factors, including different body locations, age groups, genders, and skin tones. The strong results in benchmark evaluations and real-world clinical scenarios suggest that PanDerm could enhance the management of skin diseases and serve as a model for developing multimodal foundation models in other medical specialties, potentially accelerating the integration of AI support in healthcare.
Dermatologist-like explainable AI enhances trust and confidence in diagnosing melanoma
Mar 17, 2023Abstract:Although artificial intelligence (AI) systems have been shown to improve the accuracy of initial melanoma diagnosis, the lack of transparency in how these systems identify melanoma poses severe obstacles to user acceptance. Explainable artificial intelligence (XAI) methods can help to increase transparency, but most XAI methods are unable to produce precisely located domain-specific explanations, making the explanations difficult to interpret. Moreover, the impact of XAI methods on dermatologists has not yet been evaluated. Extending on two existing classifiers, we developed an XAI system that produces text and region based explanations that are easily interpretable by dermatologists alongside its differential diagnoses of melanomas and nevi. To evaluate this system, we conducted a three-part reader study to assess its impact on clinicians' diagnostic accuracy, confidence, and trust in the XAI-support. We showed that our XAI's explanations were highly aligned with clinicians' explanations and that both the clinicians' trust in the support system and their confidence in their diagnoses were significantly increased when using our XAI compared to using a conventional AI system. The clinicians' diagnostic accuracy was numerically, albeit not significantly, increased. This work demonstrates that clinicians are willing to adopt such an XAI system, motivating their future use in the clinic.
A Patient-Centric Dataset of Images and Metadata for Identifying Melanomas Using Clinical Context
Aug 07, 2020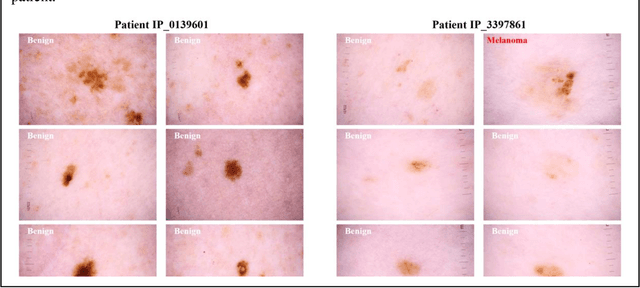
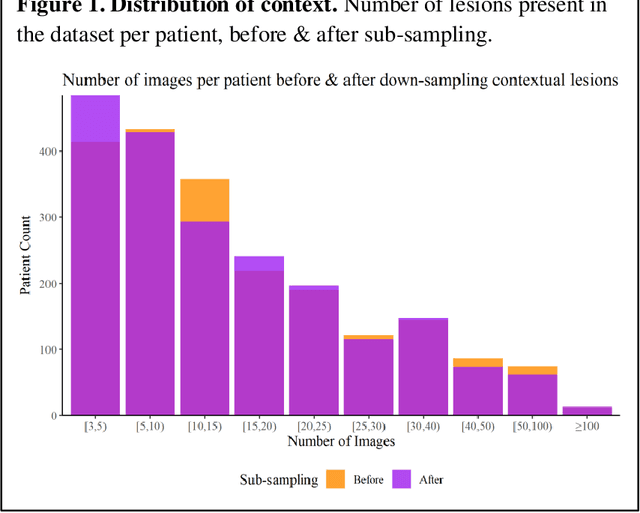
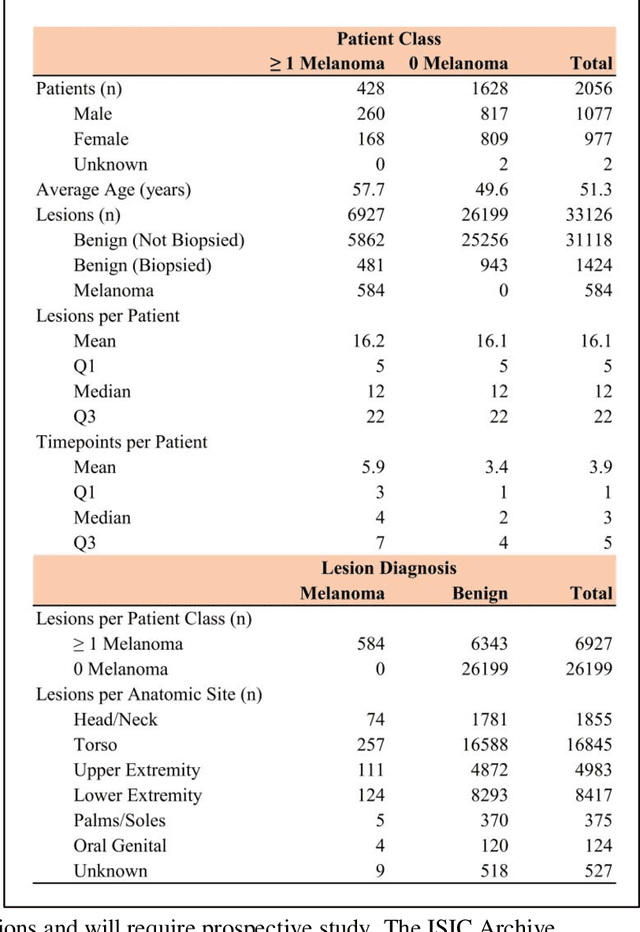
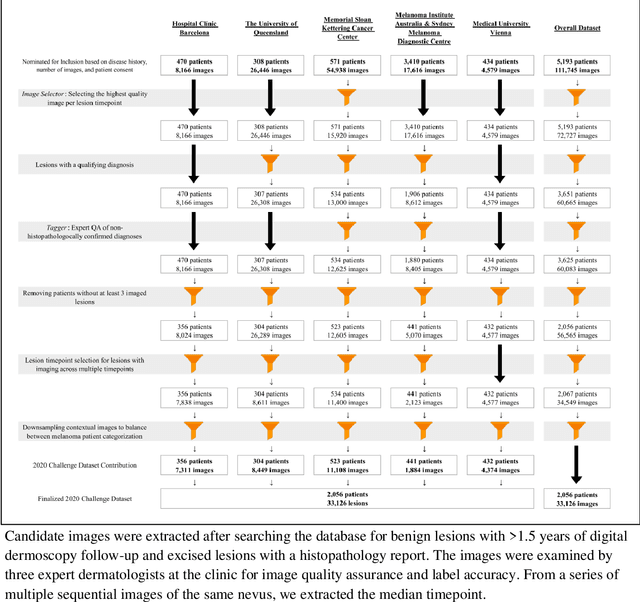
Abstract:Prior skin image datasets have not addressed patient-level information obtained from multiple skin lesions from the same patient. Though artificial intelligence classification algorithms have achieved expert-level performance in controlled studies examining single images, in practice dermatologists base their judgment holistically from multiple lesions on the same patient. The 2020 SIIM-ISIC Melanoma Classification challenge dataset described herein was constructed to address this discrepancy between prior challenges and clinical practice, providing for each image in the dataset an identifier allowing lesions from the same patient to be mapped to one another. This patient-level contextual information is frequently used by clinicians to diagnose melanoma and is especially useful in ruling out false positives in patients with many atypical nevi. The dataset represents 2,056 patients from three continents with an average of 16 lesions per patient, consisting of 33,126 dermoscopic images and 584 histopathologically confirmed melanomas compared with benign melanoma mimickers.
Detecting cutaneous basal cell carcinomas in ultra-high resolution and weakly labelled histopathological images
Dec 02, 2019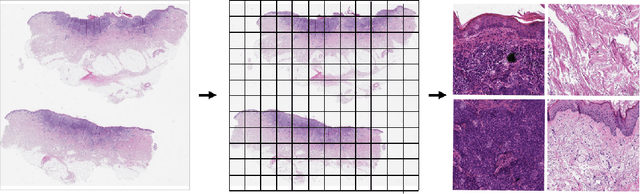

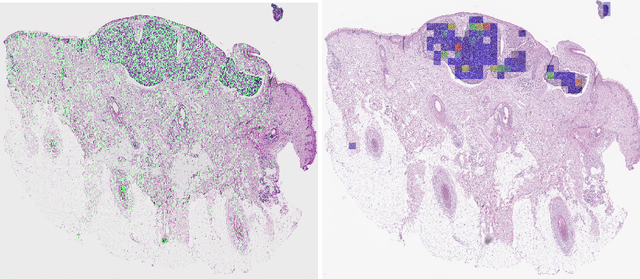
Abstract:Diagnosing basal cell carcinomas (BCC), one of the most common cutaneous malignancies in humans, is a task regularly performed by pathologists and dermato-pathologists. Improving histological diagnosis by providing diagnosis suggestions, i.e. computer-assisted diagnoses is actively researched to improve safety, quality and efficiency. Increasingly, machine learning methods are applied due to their superior performance. However, typical images obtained by scanning histological sections often have a resolution that is prohibitive for processing with current state-of-the-art neural networks. Furthermore, the data pose a problem of weak labels, since only a tiny fraction of the image is indicative of the disease class, whereas a large fraction of the image is highly similar to the non-disease class. The aim of this study is to evaluate whether it is possible to detect basal cell carcinomas in histological sections using attention-based deep learning models and to overcome the ultra-high resolution and the weak labels of whole slide images. We demonstrate that attention-based models can indeed yield almost perfect classification performance with an AUC of 0.99.
Skin Lesion Analysis Toward Melanoma Detection 2018: A Challenge Hosted by the International Skin Imaging Collaboration (ISIC)
Mar 29, 2019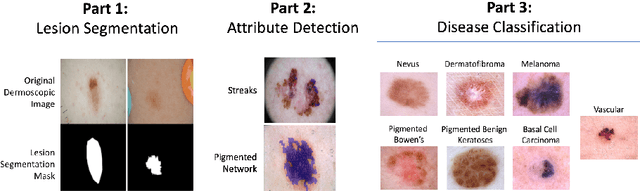
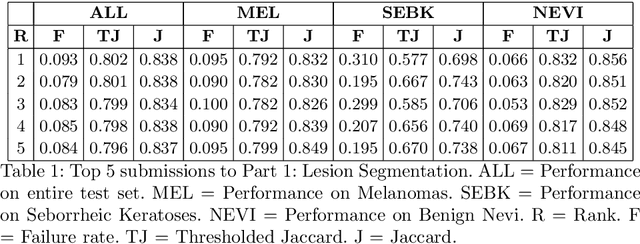
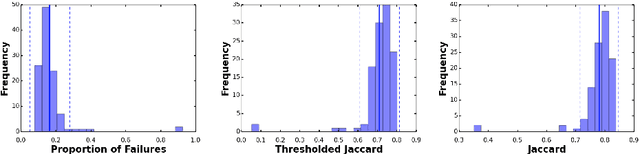
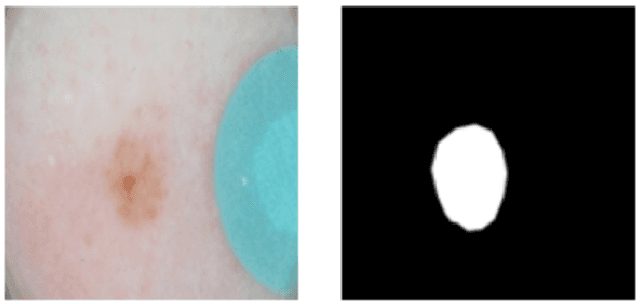
Abstract:This work summarizes the results of the largest skin image analysis challenge in the world, hosted by the International Skin Imaging Collaboration (ISIC), a global partnership that has organized the world's largest public repository of dermoscopic images of skin. The challenge was hosted in 2018 at the Medical Image Computing and Computer Assisted Intervention (MICCAI) conference in Granada, Spain. The dataset included over 12,500 images across 3 tasks. 900 users registered for data download, 115 submitted to the lesion segmentation task, 25 submitted to the lesion attribute detection task, and 159 submitted to the disease classification task. Novel evaluation protocols were established, including a new test for segmentation algorithm performance, and a test for algorithm ability to generalize. Results show that top segmentation algorithms still fail on over 10% of images on average, and algorithms with equal performance on test data can have different abilities to generalize. This is an important consideration for agencies regulating the growing set of machine learning tools in the healthcare domain, and sets a new standard for future public challenges in healthcare.
The HAM10000 Dataset: A Large Collection of Multi-Source Dermatoscopic Images of Common Pigmented Skin Lesions
Apr 02, 2018



Abstract:Training of neural networks for automated diagnosis of pigmented skin lesions is hampered by the small size and lack of diversity of available datasets of dermatoscopic images. We tackle this problem by releasing the HAM10000 ("Human Against Machine with 10000 training images") dataset. We collected dermatoscopic images from different populations acquired and stored by different modalities. Given this diversity we had to apply different acquisition and cleaning methods and developed semi-automatic workflows utilizing specifically trained neural networks. The final dataset consists of 11788 dermatoscopic images, of which 10010 will be released as a training set for academic machine learning purposes and will be publicly available through the ISIC archive. This benchmark dataset can be used for machine learning and for comparisons with human experts. Cases include a representative collection of all important diagnostic categories in the realm of pigmented lesions. More than 50% of lesions have been confirmed by pathology, the ground truth for the rest of the cases was either follow-up, expert consensus, or confirmation by in-vivo confocal microscopy.
Skin Lesion Analysis Toward Melanoma Detection: A Challenge at the 2017 International Symposium on Biomedical Imaging , Hosted by the International Skin Imaging Collaboration
Jan 08, 2018
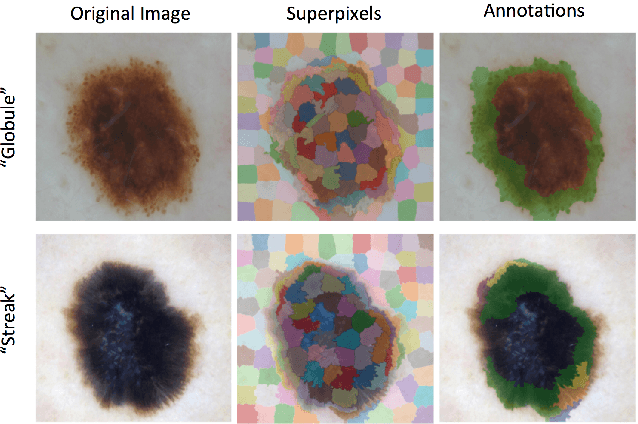
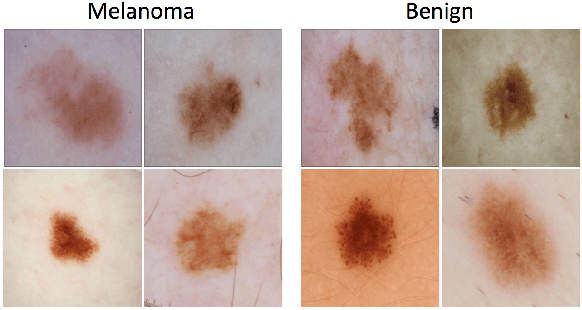
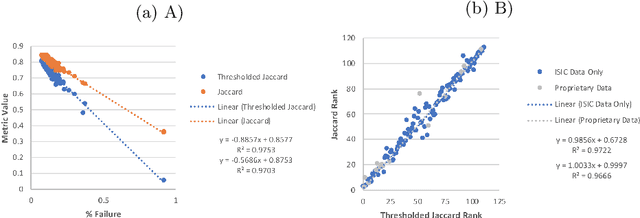
Abstract:This article describes the design, implementation, and results of the latest installment of the dermoscopic image analysis benchmark challenge. The goal is to support research and development of algorithms for automated diagnosis of melanoma, the most lethal skin cancer. The challenge was divided into 3 tasks: lesion segmentation, feature detection, and disease classification. Participation involved 593 registrations, 81 pre-submissions, 46 finalized submissions (including a 4-page manuscript), and approximately 50 attendees, making this the largest standardized and comparative study in this field to date. While the official challenge duration and ranking of participants has concluded, the dataset snapshots remain available for further research and development.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge