Guillaume Vray
ReservoirTTA: Prolonged Test-time Adaptation for Evolving and Recurring Domains
May 20, 2025Abstract:This paper introduces ReservoirTTA, a novel plug-in framework designed for prolonged test-time adaptation (TTA) in scenarios where the test domain continuously shifts over time, including cases where domains recur or evolve gradually. At its core, ReservoirTTA maintains a reservoir of domain-specialized models -- an adaptive test-time model ensemble -- that both detects new domains via online clustering over style features of incoming samples and routes each sample to the appropriate specialized model, and thereby enables domain-specific adaptation. This multi-model strategy overcomes key limitations of single model adaptation, such as catastrophic forgetting, inter-domain interference, and error accumulation, ensuring robust and stable performance on sustained non-stationary test distributions. Our theoretical analysis reveals key components that bound parameter variance and prevent model collapse, while our plug-in TTA module mitigates catastrophic forgetting of previously encountered domains. Extensive experiments on the classification corruption benchmarks, including ImageNet-C and CIFAR-10/100-C, as well as the Cityscapes$\rightarrow$ACDC semantic segmentation task, covering recurring and continuously evolving domain shifts, demonstrate that ReservoirTTA significantly improves adaptation accuracy and maintains stable performance across prolonged, recurring shifts, outperforming state-of-the-art methods.
Slide-Level Prompt Learning with Vision Language Models for Few-Shot Multiple Instance Learning in Histopathology
Mar 21, 2025Abstract:In this paper, we address the challenge of few-shot classification in histopathology whole slide images (WSIs) by utilizing foundational vision-language models (VLMs) and slide-level prompt learning. Given the gigapixel scale of WSIs, conventional multiple instance learning (MIL) methods rely on aggregation functions to derive slide-level (bag-level) predictions from patch representations, which require extensive bag-level labels for training. In contrast, VLM-based approaches excel at aligning visual embeddings of patches with candidate class text prompts but lack essential pathological prior knowledge. Our method distinguishes itself by utilizing pathological prior knowledge from language models to identify crucial local tissue types (patches) for WSI classification, integrating this within a VLM-based MIL framework. Our approach effectively aligns patch images with tissue types, and we fine-tune our model via prompt learning using only a few labeled WSIs per category. Experimentation on real-world pathological WSI datasets and ablation studies highlight our method's superior performance over existing MIL- and VLM-based methods in few-shot WSI classification tasks. Our code is publicly available at https://github.com/LTS5/SLIP.
Un-Mixing Test-Time Normalization Statistics: Combatting Label Temporal Correlation
Jan 16, 2024



Abstract:In an era where test-time adaptation methods increasingly rely on the nuanced manipulation of batch normalization (BN) parameters, one critical assumption often goes overlooked: that of independently and identically distributed (i.i.d.) test batches with respect to unknown labels. This assumption culminates in biased estimates of BN statistics and jeopardizes system stability under non-i.i.d. conditions. This paper pioneers a departure from the i.i.d. paradigm by introducing a groundbreaking strategy termed "Un-Mixing Test-Time Normalization Statistics" (UnMix-TNS). UnMix-TNS re-calibrates the instance-wise statistics used to normalize each instance in a batch by mixing it with multiple unmixed statistics components, thus inherently simulating the i.i.d. environment. The key lies in our innovative online unmixing procedure, which persistently refines these statistics components by drawing upon the closest instances from an incoming test batch. Remarkably generic in its design, UnMix-TNS seamlessly integrates with an array of state-of-the-art test-time adaptation methods and pre-trained architectures equipped with BN layers. Empirical evaluations corroborate the robustness of UnMix-TNS under varied scenarios ranging from single to continual and mixed domain shifts. UnMix-TNS stands out when handling test data streams with temporal correlation, including those with corrupted real-world non-i.i.d. streams, sustaining its efficacy even with minimal batch sizes and individual samples. Our results set a new standard for test-time adaptation, demonstrating significant improvements in both stability and performance across multiple benchmarks.
Source-Free Open-Set Domain Adaptation for Histopathological Images via Distilling Self-Supervised Vision Transformer
Jul 10, 2023



Abstract:There is a strong incentive to develop computational pathology models to i) ease the burden of tissue typology annotation from whole slide histological images; ii) transfer knowledge, e.g., tissue class separability from the withheld source domain to the distributionally shifted unlabeled target domain, and simultaneously iii) detect Open Set samples, i.e., unseen novel categories not present in the training source domain. This paper proposes a highly practical setting by addressing the abovementioned challenges in one fell swoop, i.e., source-free Open Set domain adaptation (SF-OSDA), which addresses the situation where a model pre-trained on the inaccessible source dataset can be adapted on the unlabeled target dataset containing Open Set samples. The central tenet of our proposed method is distilling knowledge from a self-supervised vision transformer trained in the target domain. We propose a novel style-based data augmentation used as hard positives for self-training a vision transformer in the target domain, yielding strongly contextualized embedding. Subsequently, semantically similar target images are clustered while the source model provides their corresponding weak pseudo-labels with unreliable confidence. Furthermore, we propose cluster relative maximum logit score (CRMLS) to rectify the confidence of the weak pseudo-labels and compute weighted class prototypes in the contextualized embedding space that are utilized for adapting the source model on the target domain. Our method significantly outperforms the previous methods, including open set detection, test-time adaptation, and SF-OSDA methods, setting the new state-of-the-art on three public histopathological datasets of colorectal cancer (CRC) assessment- Kather-16, Kather-19, and CRCTP. Our code is available at https://github.com/LTS5/Proto-SF-OSDA.
TeSLA: Test-Time Self-Learning With Automatic Adversarial Augmentation
Mar 17, 2023Abstract:Most recent test-time adaptation methods focus on only classification tasks, use specialized network architectures, destroy model calibration or rely on lightweight information from the source domain. To tackle these issues, this paper proposes a novel Test-time Self-Learning method with automatic Adversarial augmentation dubbed TeSLA for adapting a pre-trained source model to the unlabeled streaming test data. In contrast to conventional self-learning methods based on cross-entropy, we introduce a new test-time loss function through an implicitly tight connection with the mutual information and online knowledge distillation. Furthermore, we propose a learnable efficient adversarial augmentation module that further enhances online knowledge distillation by simulating high entropy augmented images. Our method achieves state-of-the-art classification and segmentation results on several benchmarks and types of domain shifts, particularly on challenging measurement shifts of medical images. TeSLA also benefits from several desirable properties compared to competing methods in terms of calibration, uncertainty metrics, insensitivity to model architectures, and source training strategies, all supported by extensive ablations. Our code and models are available on GitHub.
Self-Supervised Generative Style Transfer for One-Shot Medical Image Segmentation
Oct 05, 2021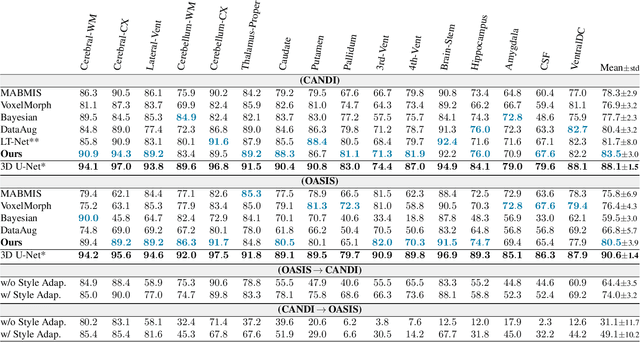
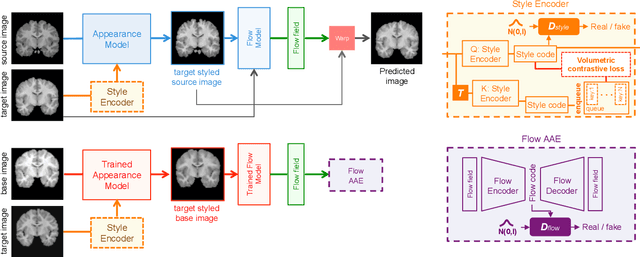
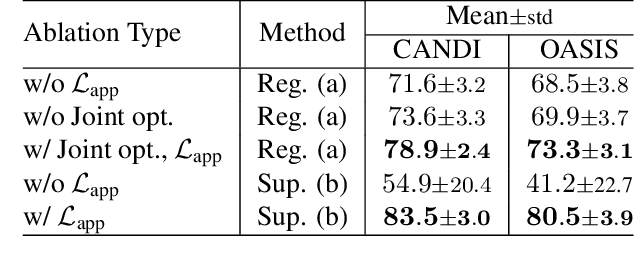
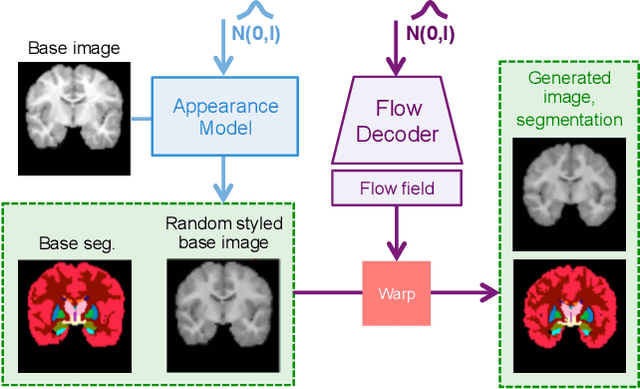
Abstract:In medical image segmentation, supervised deep networks' success comes at the cost of requiring abundant labeled data. While asking domain experts to annotate only one or a few of the cohort's images is feasible, annotating all available images is impractical. This issue is further exacerbated when pre-trained deep networks are exposed to a new image dataset from an unfamiliar distribution. Using available open-source data for ad-hoc transfer learning or hand-tuned techniques for data augmentation only provides suboptimal solutions. Motivated by atlas-based segmentation, we propose a novel volumetric self-supervised learning for data augmentation capable of synthesizing volumetric image-segmentation pairs via learning transformations from a single labeled atlas to the unlabeled data. Our work's central tenet benefits from a combined view of one-shot generative learning and the proposed self-supervised training strategy that cluster unlabeled volumetric images with similar styles together. Unlike previous methods, our method does not require input volumes at inference time to synthesize new images. Instead, it can generate diversified volumetric image-segmentation pairs from a prior distribution given a single or multi-site dataset. Augmented data generated by our method used to train the segmentation network provide significant improvements over state-of-the-art deep one-shot learning methods on the task of brain MRI segmentation. Ablation studies further exemplified that the proposed appearance model and joint training are crucial to synthesize realistic examples compared to existing medical registration methods. The code, data, and models are available at https://github.com/devavratTomar/SST.
Self-Attentive Spatial Adaptive Normalization for Cross-Modality Domain Adaptation
Mar 05, 2021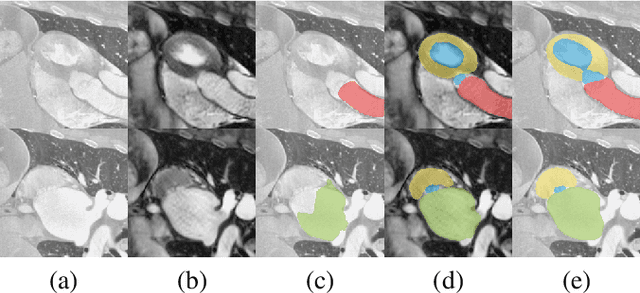
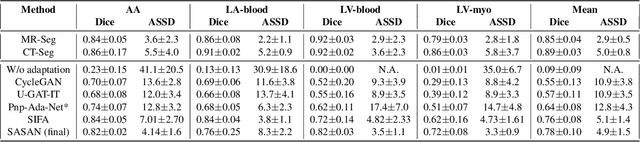
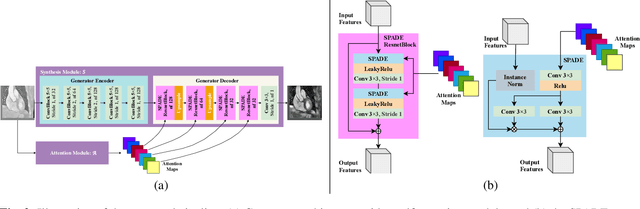
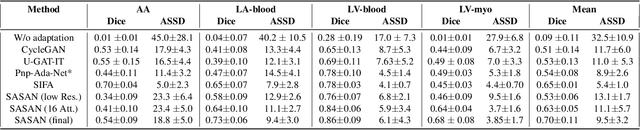
Abstract:Despite the successes of deep neural networks on many challenging vision tasks, they often fail to generalize to new test domains that are not distributed identically to the training data. The domain adaptation becomes more challenging for cross-modality medical data with a notable domain shift. Given that specific annotated imaging modalities may not be accessible nor complete. Our proposed solution is based on the cross-modality synthesis of medical images to reduce the costly annotation burden by radiologists and bridge the domain gap in radiological images. We present a novel approach for image-to-image translation in medical images, capable of supervised or unsupervised (unpaired image data) setups. Built upon adversarial training, we propose a learnable self-attentive spatial normalization of the deep convolutional generator network's intermediate activations. Unlike previous attention-based image-to-image translation approaches, which are either domain-specific or require distortion of the source domain's structures, we unearth the importance of the auxiliary semantic information to handle the geometric changes and preserve anatomical structures during image translation. We achieve superior results for cross-modality segmentation between unpaired MRI and CT data for multi-modality whole heart and multi-modal brain tumor MRI (T1/T2) datasets compared to the state-of-the-art methods. We also observe encouraging results in cross-modality conversion for paired MRI and CT images on a brain dataset. Furthermore, a detailed analysis of the cross-modality image translation, thorough ablation studies confirm our proposed method's efficacy.
Anomaly Detection on X-Rays Using Self-Supervised Aggregation Learning
Oct 19, 2020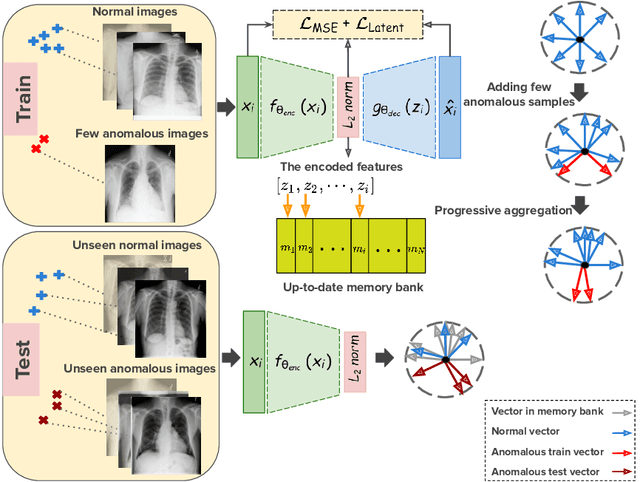
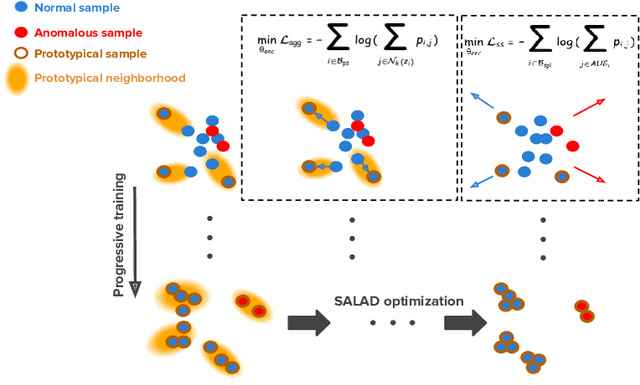
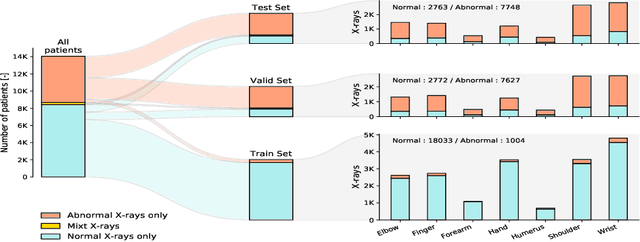
Abstract:Deep anomaly detection models using a supervised mode of learning usually work under a closed set assumption and suffer from overfitting to previously seen rare anomalies at training, which hinders their applicability in a real scenario. In addition, obtaining annotations for X-rays is very time consuming and requires extensive training of radiologists. Hence, training anomaly detection in a fully unsupervised or self-supervised fashion would be advantageous, allowing a significant reduction of time spent on the report by radiologists. In this paper, we present SALAD, an end-to-end deep self-supervised methodology for anomaly detection on X-Ray images. The proposed method is based on an optimization strategy in which a deep neural network is encouraged to represent prototypical local patterns of the normal data in the embedding space. During training, we record the prototypical patterns of normal training samples via a memory bank. Our anomaly score is then derived by measuring similarity to a weighted combination of normal prototypical patterns within a memory bank without using any anomalous patterns. We present extensive experiments on the challenging NIH Chest X-rays and MURA dataset, which indicate that our algorithm improves state-of-the-art methods by a wide margin.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge