Paul Bonnington
A General-Purpose Multimodal Foundation Model for Dermatology
Oct 19, 2024



Abstract:Diagnosing and treating skin diseases require advanced visual skills across multiple domains and the ability to synthesize information from various imaging modalities. Current deep learning models, while effective at specific tasks such as diagnosing skin cancer from dermoscopic images, fall short in addressing the complex, multimodal demands of clinical practice. Here, we introduce PanDerm, a multimodal dermatology foundation model pretrained through self-supervised learning on a dataset of over 2 million real-world images of skin diseases, sourced from 11 clinical institutions across 4 imaging modalities. We evaluated PanDerm on 28 diverse datasets covering a range of clinical tasks, including skin cancer screening, phenotype assessment and risk stratification, diagnosis of neoplastic and inflammatory skin diseases, skin lesion segmentation, change monitoring, and metastasis prediction and prognosis. PanDerm achieved state-of-the-art performance across all evaluated tasks, often outperforming existing models even when using only 5-10% of labeled data. PanDerm's clinical utility was demonstrated through reader studies in real-world clinical settings across multiple imaging modalities. It outperformed clinicians by 10.2% in early-stage melanoma detection accuracy and enhanced clinicians' multiclass skin cancer diagnostic accuracy by 11% in a collaborative human-AI setting. Additionally, PanDerm demonstrated robust performance across diverse demographic factors, including different body locations, age groups, genders, and skin tones. The strong results in benchmark evaluations and real-world clinical scenarios suggest that PanDerm could enhance the management of skin diseases and serve as a model for developing multimodal foundation models in other medical specialties, potentially accelerating the integration of AI support in healthcare.
Revamping AI Models in Dermatology: Overcoming Critical Challenges for Enhanced Skin Lesion Diagnosis
Nov 02, 2023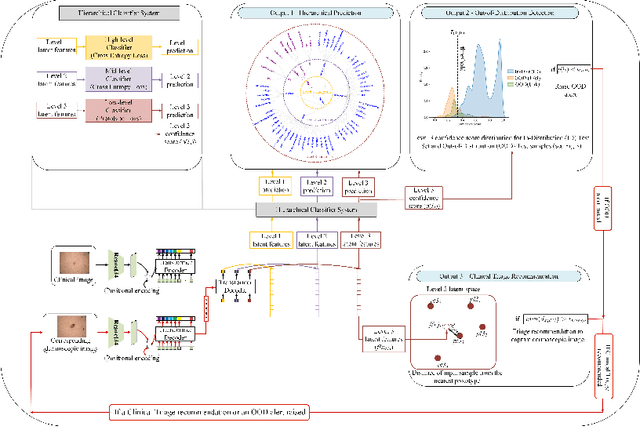
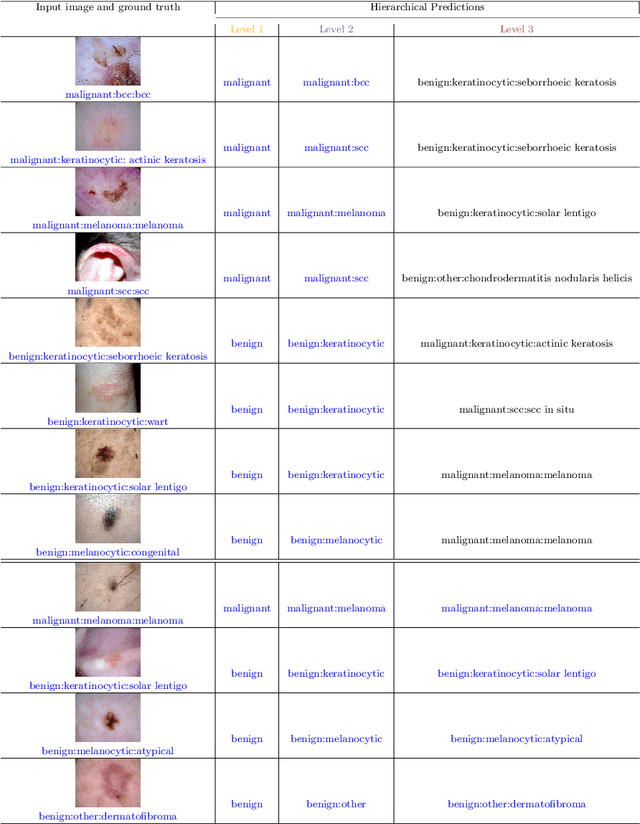

Abstract:The surge in developing deep learning models for diagnosing skin lesions through image analysis is notable, yet their clinical black faces challenges. Current dermatology AI models have limitations: limited number of possible diagnostic outputs, lack of real-world testing on uncommon skin lesions, inability to detect out-of-distribution images, and over-reliance on dermoscopic images. To address these, we present an All-In-One \textbf{H}ierarchical-\textbf{O}ut of Distribution-\textbf{C}linical Triage (HOT) model. For a clinical image, our model generates three outputs: a hierarchical prediction, an alert for out-of-distribution images, and a recommendation for dermoscopy if clinical image alone is insufficient for diagnosis. When the recommendation is pursued, it integrates both clinical and dermoscopic images to deliver final diagnosis. Extensive experiments on a representative cutaneous lesion dataset demonstrate the effectiveness and synergy of each component within our framework. Our versatile model provides valuable decision support for lesion diagnosis and sets a promising precedent for medical AI applications.
Skin Lesion Recognition with Class-Hierarchy Regularized Hyperbolic Embeddings
Sep 13, 2022Abstract:In practice, many medical datasets have an underlying taxonomy defined over the disease label space. However, existing classification algorithms for medical diagnoses often assume semantically independent labels. In this study, we aim to leverage class hierarchy with deep learning algorithms for more accurate and reliable skin lesion recognition. We propose a hyperbolic network to learn image embeddings and class prototypes jointly. The hyperbola provably provides a space for modeling hierarchical relations better than Euclidean geometry. Meanwhile, we restrict the distribution of hyperbolic prototypes with a distance matrix that is encoded from the class hierarchy. Accordingly, the learned prototypes preserve the semantic class relations in the embedding space and we can predict the label of an image by assigning its feature to the nearest hyperbolic class prototype. We use an in-house skin lesion dataset which consists of around 230k dermoscopic images on 65 skin diseases to verify our method. Extensive experiments provide evidence that our model can achieve higher accuracy with less severe classification errors than models without considering class relations.
Out-of-Distribution Detection for Long-tailed and Fine-grained Skin Lesion Images
Jun 30, 2022
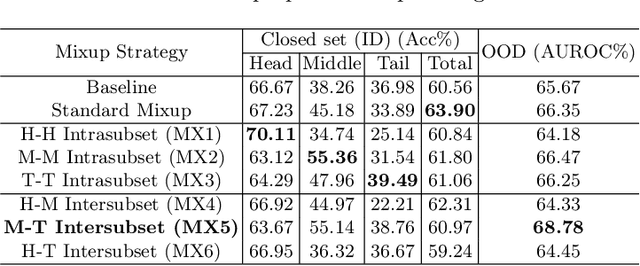

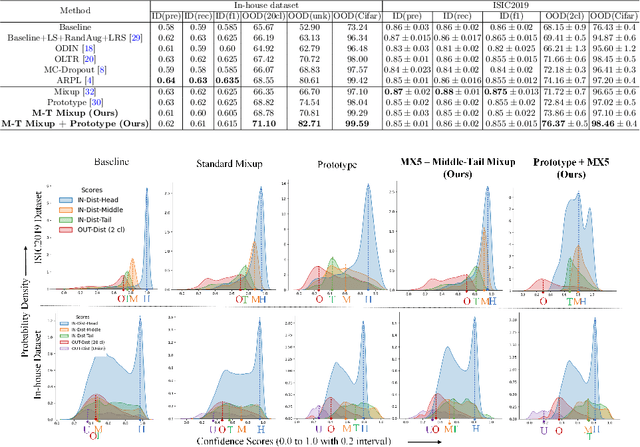
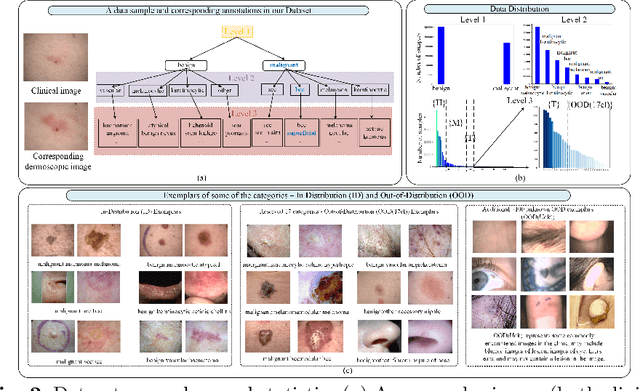
Abstract:Recent years have witnessed a rapid development of automated methods for skin lesion diagnosis and classification. Due to an increasing deployment of such systems in clinics, it has become important to develop a more robust system towards various Out-of-Distribution(OOD) samples (unknown skin lesions and conditions). However, the current deep learning models trained for skin lesion classification tend to classify these OOD samples incorrectly into one of their learned skin lesion categories. To address this issue, we propose a simple yet strategic approach that improves the OOD detection performance while maintaining the multi-class classification accuracy for the known categories of skin lesion. To specify, this approach is built upon a realistic scenario of a long-tailed and fine-grained OOD detection task for skin lesion images. Through this approach, 1) First, we target the mixup amongst middle and tail classes to address the long-tail problem. 2) Later, we combine the above mixup strategy with prototype learning to address the fine-grained nature of the dataset. The unique contribution of this paper is two-fold, justified by extensive experiments. First, we present a realistic problem setting of OOD task for skin lesion. Second, we propose an approach to target the long-tailed and fine-grained aspects of the problem setting simultaneously to increase the OOD performance.
Flexible Sampling for Long-tailed Skin Lesion Classification
Apr 07, 2022


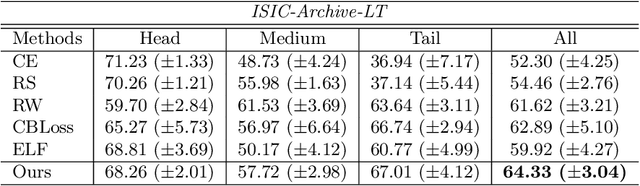
Abstract:Most of the medical tasks naturally exhibit a long-tailed distribution due to the complex patient-level conditions and the existence of rare diseases. Existing long-tailed learning methods usually treat each class equally to re-balance the long-tailed distribution. However, considering that some challenging classes may present diverse intra-class distributions, re-balancing all classes equally may lead to a significant performance drop. To address this, in this paper, we propose a curriculum learning-based framework called Flexible Sampling for the long-tailed skin lesion classification task. Specifically, we initially sample a subset of training data as anchor points based on the individual class prototypes. Then, these anchor points are used to pre-train an inference model to evaluate the per-class learning difficulty. Finally, we use a curriculum sampling module to dynamically query new samples from the rest training samples with the learning difficulty-aware sampling probability. We evaluated our model against several state-of-the-art methods on the ISIC dataset. The results with two long-tailed settings have demonstrated the superiority of our proposed training strategy, which achieves a new benchmark for long-tailed skin lesion classification.
Early Melanoma Diagnosis with Sequential Dermoscopic Images
Oct 12, 2021
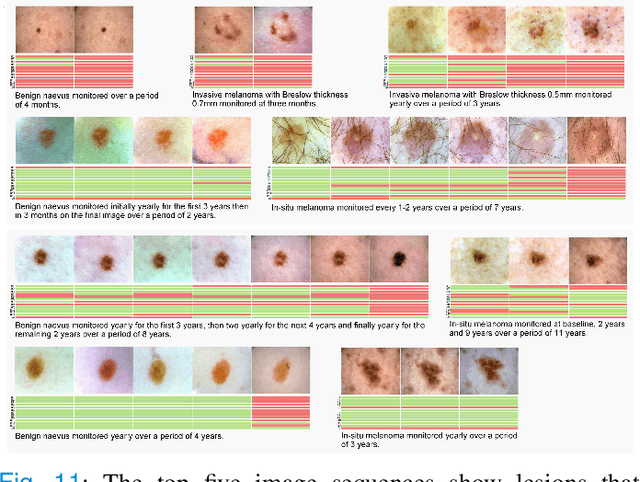
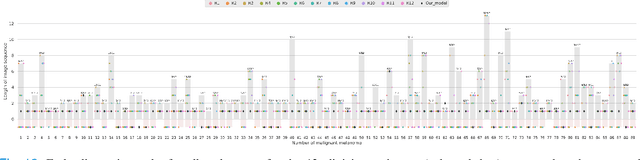
Abstract:Dermatologists often diagnose or rule out early melanoma by evaluating the follow-up dermoscopic images of skin lesions. However, existing algorithms for early melanoma diagnosis are developed using single time-point images of lesions. Ignoring the temporal, morphological changes of lesions can lead to misdiagnosis in borderline cases. In this study, we propose a framework for automated early melanoma diagnosis using sequential dermoscopic images. To this end, we construct our method in three steps. First, we align sequential dermoscopic images of skin lesions using estimated Euclidean transformations, extract the lesion growth region by computing image differences among the consecutive images, and then propose a spatio-temporal network to capture the dermoscopic changes from aligned lesion images and the corresponding difference images. Finally, we develop an early diagnosis module to compute probability scores of malignancy for lesion images over time. We collected 179 serial dermoscopic imaging data from 122 patients to verify our method. Extensive experiments show that the proposed model outperforms other commonly used sequence models. We also compared the diagnostic results of our model with those of seven experienced dermatologists and five registrars. Our model achieved higher diagnostic accuracy than clinicians (63.69% vs. 54.33%, respectively) and provided an earlier diagnosis of melanoma (60.7% vs. 32.7% of melanoma correctly diagnosed on the first follow-up images). These results demonstrate that our model can be used to identify melanocytic lesions that are at high-risk of malignant transformation earlier in the disease process and thereby redefine what is possible in the early detection of melanoma.
Leveraging Regular Fundus Images for Training UWF Fundus Diagnosis Models via Adversarial Learning and Pseudo-Labeling
Nov 27, 2020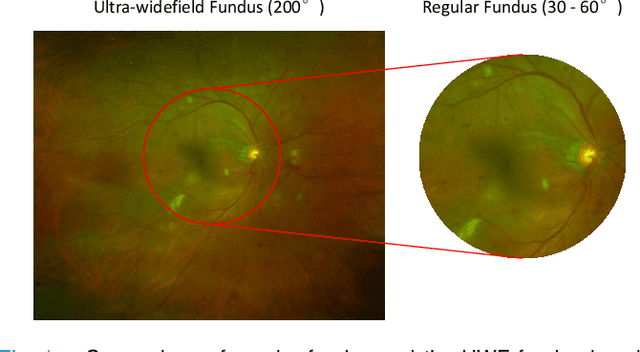


Abstract:Recently, ultra-widefield (UWF) 200-degree fundus imaging by Optos cameras has gradually been introduced because of its broader insights for detecting more information on the fundus than regular 30-degree - 60-degree fundus cameras. Compared with UWF fundus images, regular fundus images contain a large amount of high-quality and well-annotated data. Due to the domain gap, models trained by regular fundus images to recognize UWF fundus images perform poorly. Hence, given that annotating medical data is labor intensive and time consuming, in this paper, we explore how to leverage regular fundus images to improve the limited UWF fundus data and annotations for more efficient training. We propose the use of a modified cycle generative adversarial network (CycleGAN) model to bridge the gap between regular and UWF fundus and generate additional UWF fundus images for training. A consistency regularization term is proposed in the loss of the GAN to improve and regulate the quality of the generated data. Our method does not require that images from the two domains be paired or even that the semantic labels be the same, which provides great convenience for data collection. Furthermore, we show that our method is robust to noise and errors introduced by the generated unlabeled data with the pseudo-labeling technique. We evaluated the effectiveness of our methods on several common fundus diseases and tasks, such as diabetic retinopathy (DR) classification, lesion detection and tessellated fundus segmentation. The experimental results demonstrate that our proposed method simultaneously achieves superior generalizability of the learned representations and performance improvements in multiple tasks.
Synergic Adversarial Label Learning with DR and AMD for Retinal Image Grading
Mar 27, 2020
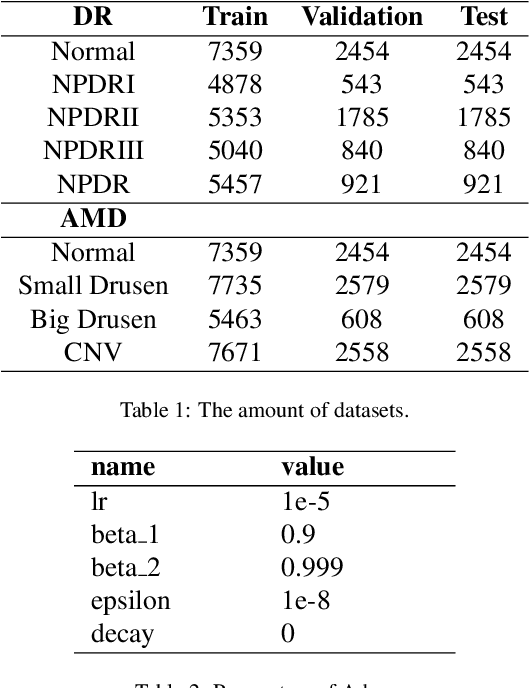

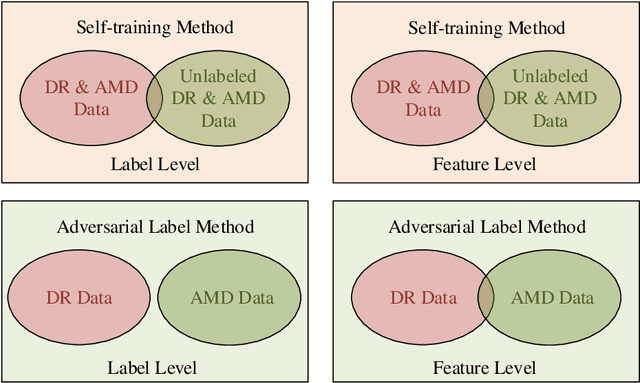
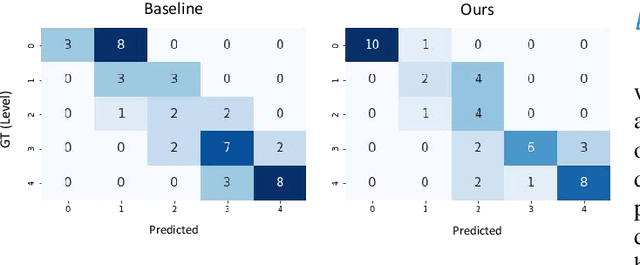
Abstract:The need for comprehensive and automated screening methods for retinal image classification has long been recognized. Well-qualified doctors annotated images are very expensive and only a limited amount of data is available for various retinal diseases such as age-related macular degeneration (AMD) and diabetic retinopathy (DR). Some studies show that AMD and DR share some common features like hemorrhagic points and exudation but most classification algorithms only train those disease models independently. Inspired by knowledge distillation where additional monitoring signals from various sources is beneficial to train a robust model with much fewer data. We propose a method called synergic adversarial label learning (SALL) which leverages relevant retinal disease labels in both semantic and feature space as additional signals and train the model in a collaborative manner. Our experiments on DR and AMD fundus image classification task demonstrate that the proposed method can significantly improve the accuracy of the model for grading diseases. In addition, we conduct additional experiments to show the effectiveness of SALL from the aspects of reliability and interpretability in the context of medical imaging application.
Bridge the Domain Gap Between Ultra-wide-field and Traditional Fundus Images via Adversarial Domain Adaptation
Mar 24, 2020

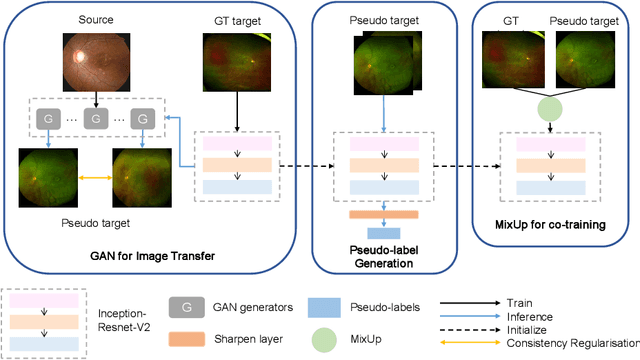
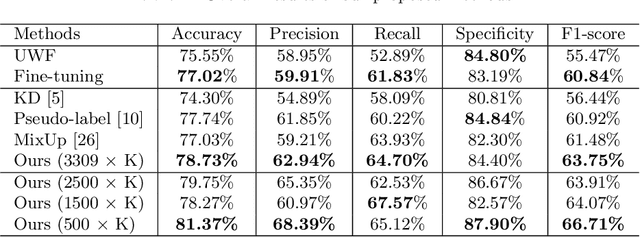
Abstract:For decades, advances in retinal imaging technology have enabled effective diagnosis and management of retinal disease using fundus cameras. Recently, ultra-wide-field (UWF) fundus imaging by Optos camera is gradually put into use because of its broader insights on fundus for some lesions that are not typically seen in traditional fundus images. Research on traditional fundus images is an active topic but studies on UWF fundus images are few. One of the most important reasons is that UWF fundus images are hard to obtain. In this paper, for the first time, we explore domain adaptation from the traditional fundus to UWF fundus images. We propose a flexible framework to bridge the domain gap between two domains and co-train a UWF fundus diagnosis model by pseudo-labelling and adversarial learning. We design a regularisation technique to regulate the domain adaptation. Also, we apply MixUp to overcome the over-fitting issue from incorrect generated pseudo-labels. Our experimental results on either single or both domains demonstrate that the proposed method can well adapt and transfer the knowledge from traditional fundus images to UWF fundus images and improve the performance of retinal disease recognition.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge