Shekhar S. Chandra
SAMRI: Segment Anything Model for MRI
Oct 30, 2025Abstract:Accurate magnetic resonance imaging (MRI) segmentation is crucial for clinical decision-making, but remains labor-intensive when performed manually. Convolutional neural network (CNN)-based methods can be accurate and efficient, but often generalize poorly to MRI's variable contrast, intensity inhomogeneity, and protocols. Although the transformer-based Segment Anything Model (SAM) has demonstrated remarkable generalizability in natural images, existing adaptations often treat MRI as another imaging modality, overlooking these modality-specific challenges. We present SAMRI, an MRI-specialized SAM trained and validated on 1.1 million labeled MR slices spanning whole-body organs and pathologies. We demonstrate that SAM can be effectively adapted to MRI by simply fine-tuning its mask decoder using a two-stage strategy, reducing training time by 94% and trainable parameters by 96% versus full-model retraining. Across diverse MRI segmentation tasks, SAMRI achieves a mean Dice of 0.87, delivering state-of-the-art accuracy across anatomical regions and robust generalization on unseen structures, particularly small and clinically important structures.
Machine Learning Applications in Traumatic Brain Injury: A Spotlight on Mild TBI
Jan 11, 2024



Abstract:Traumatic Brain Injury (TBI) poses a significant global public health challenge, contributing to high morbidity and mortality rates and placing a substantial economic burden on healthcare systems worldwide. The diagnosis of TBI relies on clinical information along with Computed Tomography (CT) scans. Addressing the multifaceted challenges posed by TBI has seen the development of innovative, data-driven approaches, for this complex condition. Particularly noteworthy is the prevalence of mild TBI (mTBI), which constitutes the majority of TBI cases where conventional methods often fall short. As such, we review the state-of-the-art Machine Learning (ML) techniques applied to clinical information and CT scans in TBI, with a particular focus on mTBI. We categorize ML applications based on their data sources, and there is a spectrum of ML techniques used to date. Most of these techniques have primarily focused on diagnosis, with relatively few attempts at predicting the prognosis. This review may serve as a source of inspiration for future research studies aimed at improving the diagnosis of TBI using data-driven approaches and standard diagnostic data.
Enhancing mTBI Diagnosis with Residual Triplet Convolutional Neural Network Using 3D CT
Nov 23, 2023Abstract:Mild Traumatic Brain Injury (mTBI) is a common and challenging condition to diagnose accurately. Timely and precise diagnosis is essential for effective treatment and improved patient outcomes. Traditional diagnostic methods for mTBI often have limitations in terms of accuracy and sensitivity. In this study, we introduce an innovative approach to enhance mTBI diagnosis using 3D Computed Tomography (CT) images and a metric learning technique trained with triplet loss. To address these challenges, we propose a Residual Triplet Convolutional Neural Network (RTCNN) model to distinguish between mTBI cases and healthy ones by embedding 3D CT scans into a feature space. The triplet loss function maximizes the margin between similar and dissimilar image pairs, optimizing feature representations. This facilitates better context placement of individual cases, aids informed decision-making, and has the potential to improve patient outcomes. Our RTCNN model shows promising performance in mTBI diagnosis, achieving an average accuracy of 94.3%, a sensitivity of 94.1%, and a specificity of 95.2%, as confirmed through a five-fold cross-validation. Importantly, when compared to the conventional Residual Convolutional Neural Network (RCNN) model, the RTCNN exhibits a significant improvement, showcasing a remarkable 22.5% increase in specificity, a notable 16.2% boost in accuracy, and an 11.3% enhancement in sensitivity. Moreover, RTCNN requires lower memory resources, making it not only highly effective but also resource-efficient in minimizing false positives while maximizing its diagnostic accuracy in distinguishing normal CT scans from mTBI cases. The quantitative performance metrics provided and utilization of occlusion sensitivity maps to visually explain the model's decision-making process further enhance the interpretability and transparency of our approach.
Single Image Compressed Sensing MRI via a Self-Supervised Deep Denoising Approach
Nov 22, 2023



Abstract:Popular methods in compressed sensing (CS) are dependent on deep learning (DL), where large amounts of data are used to train non-linear reconstruction models. However, ensuring generalisability over and access to multiple datasets is challenging to realise for real-world applications. To address these concerns, this paper proposes a single image, self-supervised (SS) CS-MRI framework that enables a joint deep and sparse regularisation of CS artefacts. The approach effectively dampens structured CS artefacts, which can be difficult to remove assuming sparse reconstruction, or relying solely on the inductive biases of CNN to produce noise-free images. Image quality is thereby improved compared to either approach alone. Metrics are evaluated using Cartesian 1D masks on a brain and knee dataset, with PSNR improving by 2-4dB on average.
Multi-scale MRI reconstruction via dilated ensemble networks
Oct 07, 2023



Abstract:As aliasing artefacts are highly structural and non-local, many MRI reconstruction networks use pooling to enlarge filter coverage and incorporate global context. However, this inadvertently impedes fine detail recovery as downsampling creates a resolution bottleneck. Moreover, real and imaginary features are commonly split into separate channels, discarding phase information particularly important to high frequency textures. In this work, we introduce an efficient multi-scale reconstruction network using dilated convolutions to preserve resolution and experiment with a complex-valued version using complex convolutions. Inspired by parallel dilated filters, multiple receptive fields are processed simultaneously with branches that see both large structural artefacts and fine local features. We also adopt dense residual connections for feature aggregation to efficiently increase scale and the deep cascade global architecture to reduce overfitting. The real-valued version of this model outperformed common reconstruction architectures as well as a state-of-the-art multi-scale network whilst being three times more efficient. The complex-valued network yielded better qualitative results when more phase information was present.
Interpretable 3D Multi-Modal Residual Convolutional Neural Network for Mild Traumatic Brain Injury Diagnosis
Sep 22, 2023Abstract:Mild Traumatic Brain Injury (mTBI) is a significant public health challenge due to its high prevalence and potential for long-term health effects. Despite Computed Tomography (CT) being the standard diagnostic tool for mTBI, it often yields normal results in mTBI patients despite symptomatic evidence. This fact underscores the complexity of accurate diagnosis. In this study, we introduce an interpretable 3D Multi-Modal Residual Convolutional Neural Network (MRCNN) for mTBI diagnostic model enhanced with Occlusion Sensitivity Maps (OSM). Our MRCNN model exhibits promising performance in mTBI diagnosis, demonstrating an average accuracy of 82.4%, sensitivity of 82.6%, and specificity of 81.6%, as validated by a five-fold cross-validation process. Notably, in comparison to the CT-based Residual Convolutional Neural Network (RCNN) model, the MRCNN shows an improvement of 4.4% in specificity and 9.0% in accuracy. We show that the OSM offers superior data-driven insights into CT images compared to the Grad-CAM approach. These results highlight the efficacy of the proposed multi-modal model in enhancing the diagnostic precision of mTBI.
Ugly Ducklings or Swans: A Tiered Quadruplet Network with Patient-Specific Mining for Improved Skin Lesion Classification
Sep 18, 2023Abstract:An ugly duckling is an obviously different skin lesion from surrounding lesions of an individual, and the ugly duckling sign is a criterion used to aid in the diagnosis of cutaneous melanoma by differentiating between highly suspicious and benign lesions. However, the appearance of pigmented lesions, can change drastically from one patient to another, resulting in difficulties in visual separation of ugly ducklings. Hence, we propose DMT-Quadruplet - a deep metric learning network to learn lesion features at two tiers - patient-level and lesion-level. We introduce a patient-specific quadruplet mining approach together with a tiered quadruplet network, to drive the network to learn more contextual information both globally and locally between the two tiers. We further incorporate a dynamic margin within the patient-specific mining to allow more useful quadruplets to be mined within individuals. Comprehensive experiments show that our proposed method outperforms traditional classifiers, achieving 54% higher sensitivity than a baseline ResNet18 CNN and 37% higher than a naive triplet network in classifying ugly duckling lesions. Visualisation of the data manifold in the metric space further illustrates that DMT-Quadruplet is capable of classifying ugly duckling lesions in both patient-specific and patient-agnostic manner successfully.
Finite Compressive Sensing
Sep 15, 2023



Abstract:This paper introduces a sparse projection matrix composed of discrete (digital) periodic lines that create a pseudo-random (p.frac) sampling scheme. Our approach enables random Cartesian sampling whilst employing deterministic and one-dimensional (1D) trajectories derived from the discrete Radon transform (DRT). Unlike radial trajectories, DRT projections can be back-projected without interpolation. Thus, we also propose a novel reconstruction method based on the exact projections of the DRT called finite Fourier reconstruction (FFR). We term this combined p.frac and FFR strategy Finite Compressive Sensing (FCS), with image recovery demonstrated on experimental and simulated data; image quality comparisons are made with Cartesian random sampling in 1D and two-dimensional (2D), as well as radial under-sampling in a more constrained experiment. Our experiments indicate FCS enables 3-5dB gain in peak signal-to-noise ratio (PSNR) for 2-, 4- and 8-fold under-sampling compared to 1D Cartesian random sampling. This paper aims to: Review common sampling strategies for compressed sensing (CS)-magnetic resonance imaging (MRI) to inform the motivation of a projective and Cartesian sampling scheme. Compare the incoherence of these sampling strategies and the proposed p.frac. Compare reconstruction quality of the sampling schemes under various reconstruction strategies to determine the suitability of p.frac for CS-MRI. It is hypothesised that because p.frac is a highly incoherent sampling scheme, that reconstructions will be of high quality compared to 1D Cartesian phase-encode under-sampling.
Application of Machine Learning in Melanoma Detection and the Identification of 'Ugly Duckling' and Suspicious Naevi: A Review
Sep 05, 2023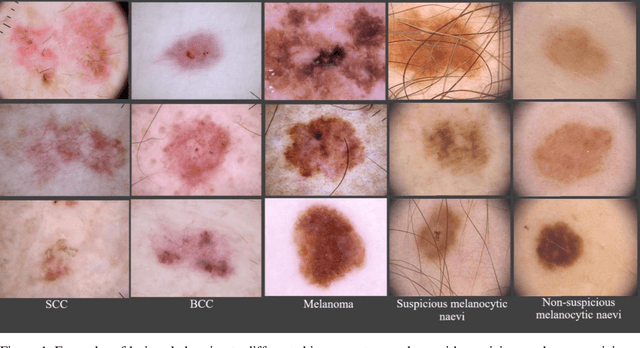
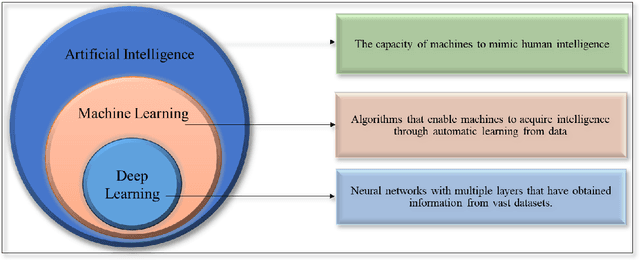

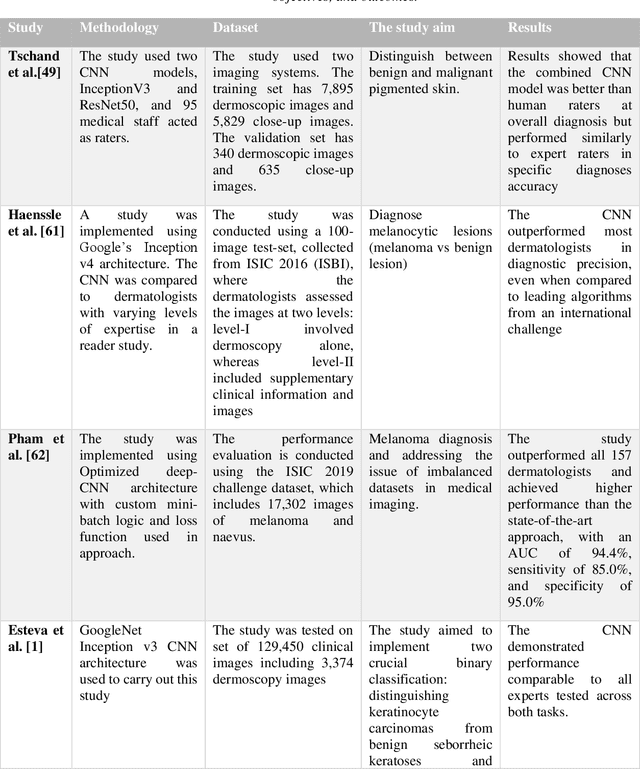
Abstract:Skin lesions known as naevi exhibit diverse characteristics such as size, shape, and colouration. The concept of an "Ugly Duckling Naevus" comes into play when monitoring for melanoma, referring to a lesion with distinctive features that sets it apart from other lesions in the vicinity. As lesions within the same individual typically share similarities and follow a predictable pattern, an ugly duckling naevus stands out as unusual and may indicate the presence of a cancerous melanoma. Computer-aided diagnosis (CAD) has become a significant player in the research and development field, as it combines machine learning techniques with a variety of patient analysis methods. Its aim is to increase accuracy and simplify decision-making, all while responding to the shortage of specialized professionals. These automated systems are especially important in skin cancer diagnosis where specialist availability is limited. As a result, their use could lead to life-saving benefits and cost reductions within healthcare. Given the drastic change in survival when comparing early stage to late-stage melanoma, early detection is vital for effective treatment and patient outcomes. Machine learning (ML) and deep learning (DL) techniques have gained popularity in skin cancer classification, effectively addressing challenges, and providing results equivalent to that of specialists. This article extensively covers modern Machine Learning and Deep Learning algorithms for detecting melanoma and suspicious naevi. It begins with general information on skin cancer and different types of naevi, then introduces AI, ML, DL, and CAD. The article then discusses the successful applications of various ML techniques like convolutional neural networks (CNN) for melanoma detection compared to dermatologists' performance. Lastly, it examines ML methods for UD naevus detection and identifying suspicious naevi.
TriFormer: A Multi-modal Transformer Framework For Mild Cognitive Impairment Conversion Prediction
Jul 14, 2023Abstract:The prediction of mild cognitive impairment (MCI) conversion to Alzheimer's disease (AD) is important for early treatment to prevent or slow the progression of AD. To accurately predict the MCI conversion to stable MCI or progressive MCI, we propose Triformer, a novel transformer-based framework with three specialized transformers to incorporate multi-model data. Triformer uses I) an image transformer to extract multi-view image features from medical scans, II) a clinical transformer to embed and correlate multi-modal clinical data, and III) a modality fusion transformer that produces an accurate prediction based on fusing the outputs from the image and clinical transformers. Triformer is evaluated on the Alzheimer's Disease Neuroimaging Initiative (ANDI)1 and ADNI2 datasets and outperforms previous state-of-the-art single and multi-modal methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge