Craig Engstrom
SAMRI: Segment Anything Model for MRI
Oct 30, 2025Abstract:Accurate magnetic resonance imaging (MRI) segmentation is crucial for clinical decision-making, but remains labor-intensive when performed manually. Convolutional neural network (CNN)-based methods can be accurate and efficient, but often generalize poorly to MRI's variable contrast, intensity inhomogeneity, and protocols. Although the transformer-based Segment Anything Model (SAM) has demonstrated remarkable generalizability in natural images, existing adaptations often treat MRI as another imaging modality, overlooking these modality-specific challenges. We present SAMRI, an MRI-specialized SAM trained and validated on 1.1 million labeled MR slices spanning whole-body organs and pathologies. We demonstrate that SAM can be effectively adapted to MRI by simply fine-tuning its mask decoder using a two-stage strategy, reducing training time by 94% and trainable parameters by 96% versus full-model retraining. Across diverse MRI segmentation tasks, SAMRI achieves a mean Dice of 0.87, delivering state-of-the-art accuracy across anatomical regions and robust generalization on unseen structures, particularly small and clinically important structures.
Evidence-aware multi-modal data fusion and its application to total knee replacement prediction
Mar 24, 2023


Abstract:Deep neural networks have been widely studied for predicting a medical condition, such as total knee replacement (TKR). It has shown that data of different modalities, such as imaging data, clinical variables and demographic information, provide complementary information and thus can improve the prediction accuracy together. However, the data sources of various modalities may not always be of high quality, and each modality may have only partial information of medical condition. Thus, predictions from different modalities can be opposite, and the final prediction may fail in the presence of such a conflict. Therefore, it is important to consider the reliability of each source data and the prediction output when making a final decision. In this paper, we propose an evidence-aware multi-modal data fusion framework based on the Dempster-Shafer theory (DST). The backbone models contain an image branch, a non-image branch and a fusion branch. For each branch, there is an evidence network that takes the extracted features as input and outputs an evidence score, which is designed to represent the reliability of the output from the current branch. The output probabilities along with the evidence scores from multiple branches are combined with the Dempster's combination rule to make a final prediction. Experimental results on the public OA initiative (OAI) dataset for the TKR prediction task show the superiority of the proposed fusion strategy on various backbone models.
Automated anomaly-aware 3D segmentation of bones and cartilages in knee MR images from the Osteoarthritis Initiative
Dec 01, 2022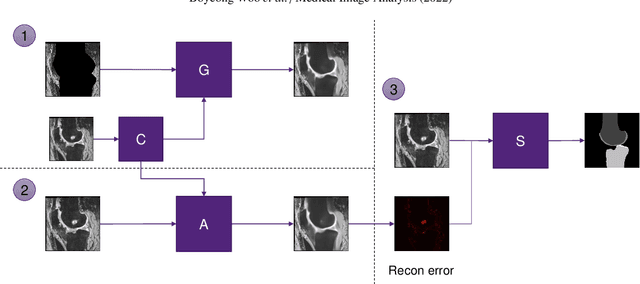
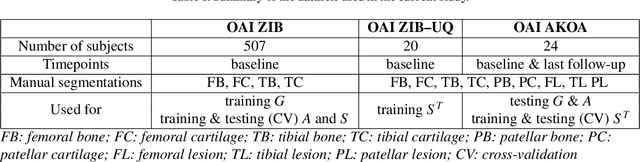
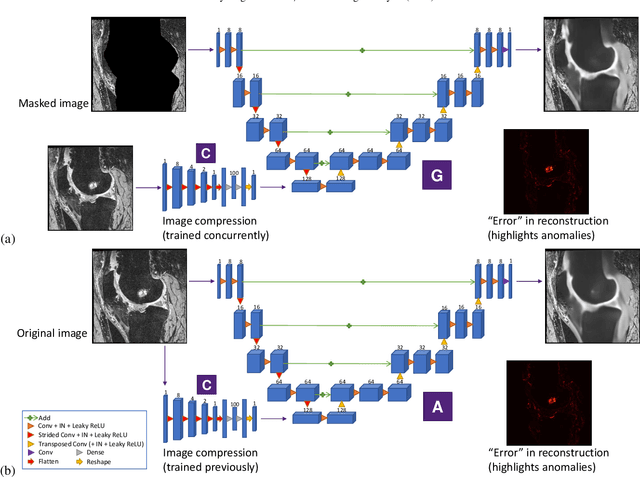
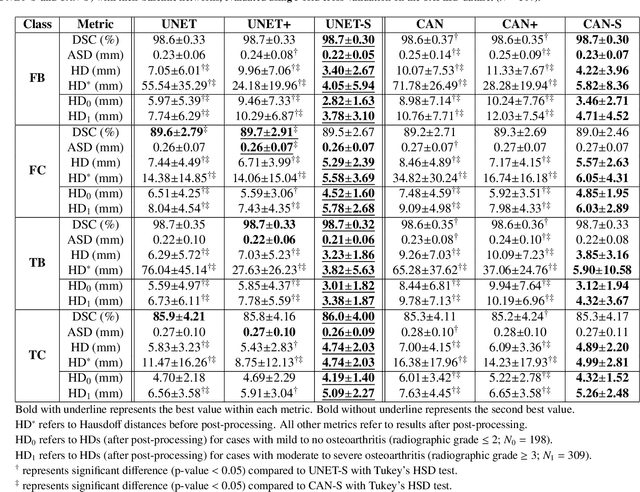
Abstract:In medical image analysis, automated segmentation of multi-component anatomical structures, which often have a spectrum of potential anomalies and pathologies, is a challenging task. In this work, we develop a multi-step approach using U-Net-based neural networks to initially detect anomalies (bone marrow lesions, bone cysts) in the distal femur, proximal tibia and patella from 3D magnetic resonance (MR) images of the knee in individuals with varying grades of osteoarthritis. Subsequently, the extracted data are used for downstream tasks involving semantic segmentation of individual bone and cartilage volumes as well as bone anomalies. For anomaly detection, the U-Net-based models were developed to reconstruct the bone profiles of the femur and tibia in images via inpainting so anomalous bone regions could be replaced with close to normal appearances. The reconstruction error was used to detect bone anomalies. A second anomaly-aware network, which was compared to anomaly-na\"ive segmentation networks, was used to provide a final automated segmentation of the femoral, tibial and patellar bones and cartilages from the knee MR images containing a spectrum of bone anomalies. The anomaly-aware segmentation approach provided up to 58% reduction in Hausdorff distances for bone segmentations compared to the results from the anomaly-na\"ive segmentation networks. In addition, the anomaly-aware networks were able to detect bone lesions in the MR images with greater sensitivity and specificity (area under the receiver operating characteristic curve [AUC] up to 0.896) compared to the anomaly-na\"ive segmentation networks (AUC up to 0.874).
Structure Guided Manifolds for Discovery of Disease Characteristics
Sep 24, 2022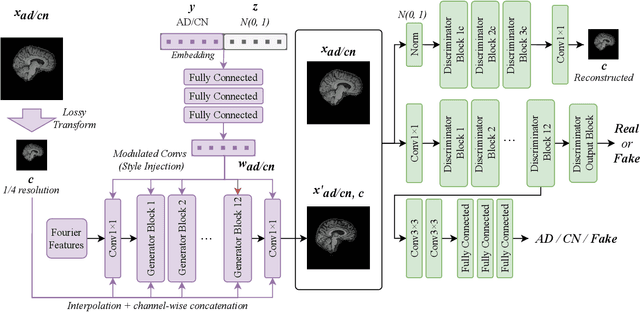
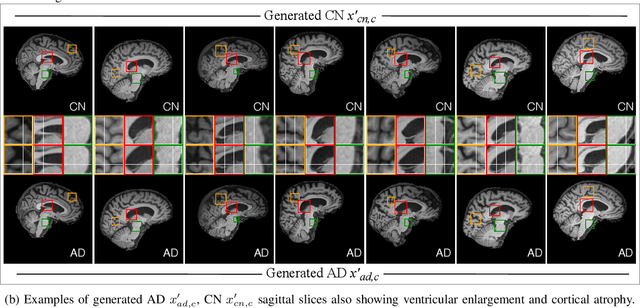
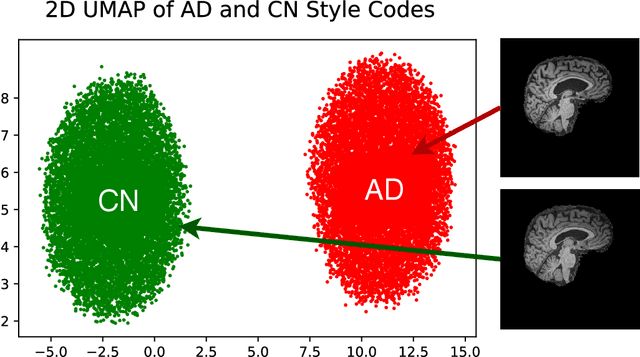

Abstract:In medical image analysis, the subtle visual characteristics of many diseases are challenging to discern, particularly due to the lack of paired data. For example, in mild Alzheimer's Disease (AD), brain tissue atrophy can be difficult to observe from pure imaging data, especially without paired AD and Cognitively Normal ( CN ) data for comparison. This work presents Disease Discovery GAN ( DiDiGAN), a weakly-supervised style-based framework for discovering and visualising subtle disease features. DiDiGAN learns a disease manifold of AD and CN visual characteristics, and the style codes sampled from this manifold are imposed onto an anatomical structural "blueprint" to synthesise paired AD and CN magnetic resonance images (MRIs). To suppress non-disease-related variations between the generated AD and CN pairs, DiDiGAN leverages a structural constraint with cycle consistency and anti-aliasing to enforce anatomical correspondence. When tested on the Alzheimer's Disease Neuroimaging Initiative ( ADNI) dataset, DiDiGAN showed key AD characteristics (reduced hippocampal volume, ventricular enlargement, and atrophy of cortical structures) through synthesising paired AD and CN scans. The qualitative results were backed up by automated brain volume analysis, where systematic pair-wise reductions in brain tissue structures were also measured
Transformer Compressed Sensing via Global Image Tokens
Mar 27, 2022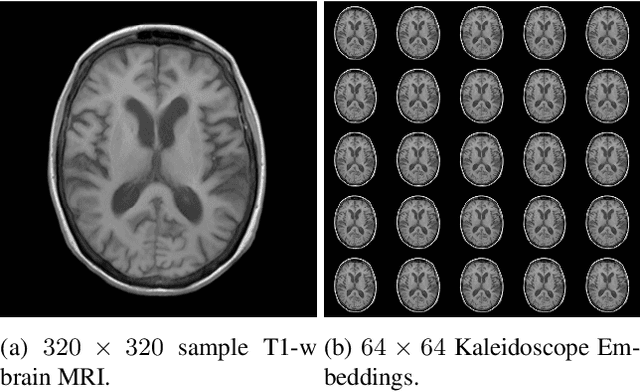

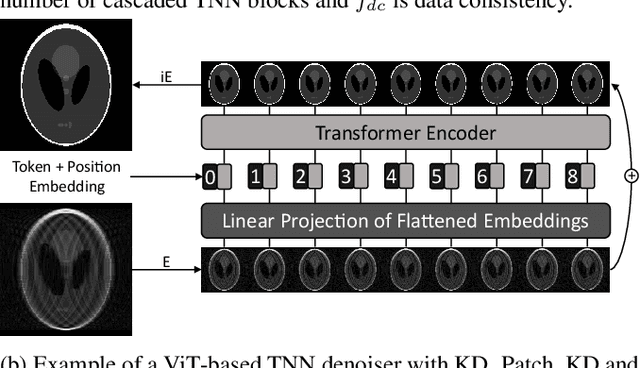
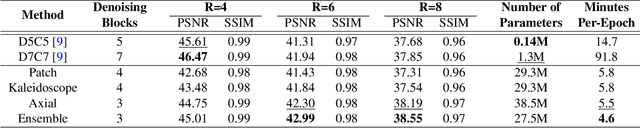
Abstract:Convolutional neural networks (CNN) have demonstrated outstanding Compressed Sensing (CS) performance compared to traditional, hand-crafted methods. However, they are broadly limited in terms of generalisability, inductive bias and difficulty to model long distance relationships. Transformer neural networks (TNN) overcome such issues by implementing an attention mechanism designed to capture dependencies between inputs. However, high-resolution tasks typically require vision Transformers (ViT) to decompose an image into patch-based tokens, limiting inputs to inherently local contexts. We propose a novel image decomposition that naturally embeds images into low-resolution inputs. These Kaleidoscope tokens (KD) provide a mechanism for global attention, at the same computational cost as a patch-based approach. To showcase this development, we replace CNN components in a well-known CS-MRI neural network with TNN blocks and demonstrate the improvements afforded by KD. We also propose an ensemble of image tokens, which enhance overall image quality and reduces model size. Supplementary material is available: https://github.com/uqmarlonbran/TCS.git
Automated volumetric and statistical shape assessment of cam-type morphology of the femoral head-neck region from 3D magnetic resonance images
Dec 06, 2021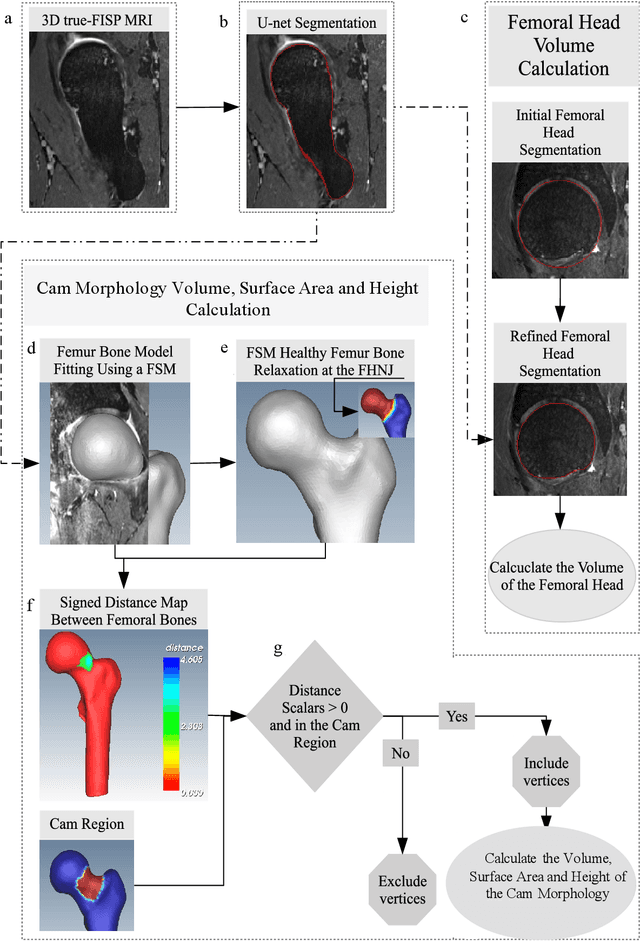
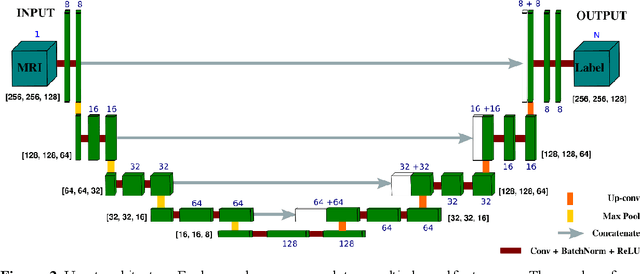

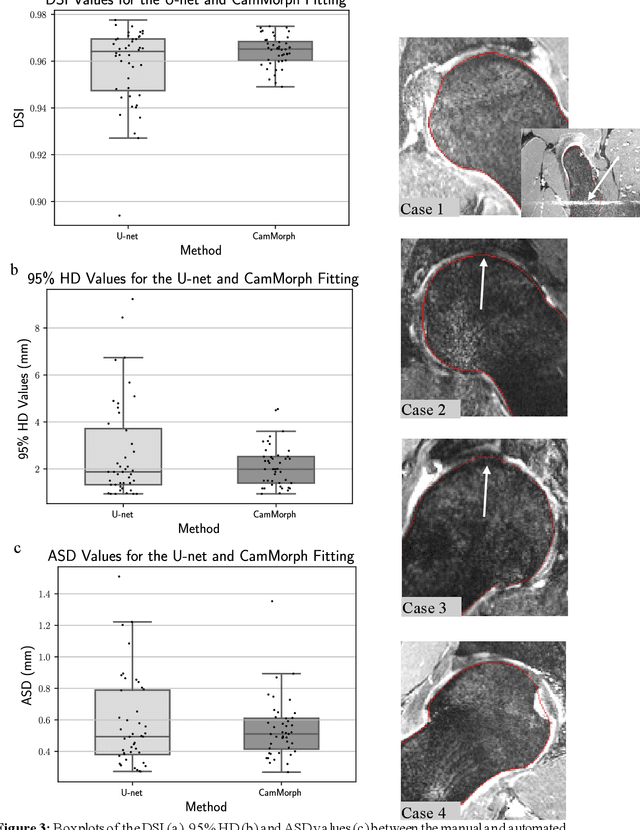
Abstract:Femoroacetabular impingement (FAI) cam morphology is routinely assessed using two-dimensional alpha angles which do not provide specific data on cam size characteristics. The purpose of this study is to implement a novel, automated three-dimensional (3D) pipeline, CamMorph, for segmentation and measurement of cam volume, surface area and height from magnetic resonance (MR) images in patients with FAI. The CamMorph pipeline involves two processes: i) proximal femur segmentation using an approach integrating 3D U-net with focused shape modelling (FSM); ii) use of patient-specific anatomical information from 3D FSM to simulate healthy femoral bone models and pathological region constraints to identify cam bone mass. Agreement between manual and automated segmentation of the proximal femur was evaluated with the Dice similarity index (DSI) and surface distance measures. Independent t-tests or Mann-Whitney U rank tests were used to compare the femoral head volume, cam volume, surface area and height data between female and male patients with FAI. There was a mean DSI value of 0.964 between manual and automated segmentation of proximal femur volume. Compared to female FAI patients, male patients had a significantly larger mean femoral head volume (66.12cm3 v 46.02cm3, p<0.001). Compared to female FAI patients, male patients had a significantly larger mean cam volume (1136.87mm3 v 337.86mm3, p<0.001), surface area (657.36mm2 v 306.93mm2 , p<0.001), maximum-height (3.89mm v 2.23mm, p<0.001) and average-height (1.94mm v 1.00mm, p<0.001). Automated analyses of 3D MR images from patients with FAI using the CamMorph pipeline showed that, in comparison with female patients, male patients had significantly greater cam volume, surface area and height.
Manipulating Medical Image Translation with Manifold Disentanglement
Nov 27, 2020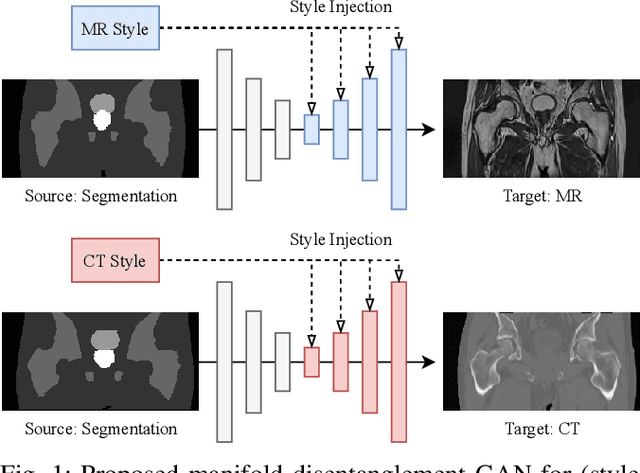
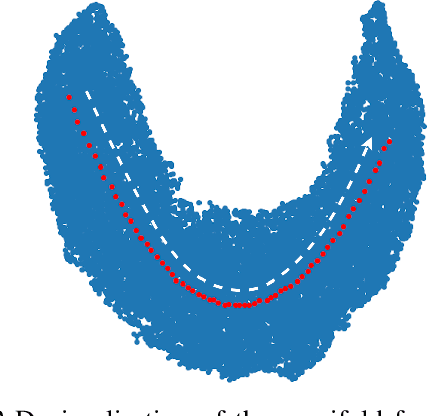

Abstract:Medical image translation (e.g. CT to MR) is a challenging task as it requires I) faithful translation of domain-invariant features (e.g. shape information of anatomical structures) and II) realistic synthesis of target-domain features (e.g. tissue appearance in MR). In this work, we propose Manifold Disentanglement Generative Adversarial Network (MDGAN), a novel image translation framework that explicitly models these two types of features. It employs a fully convolutional generator to model domain-invariant features, and it uses style codes to separately model target-domain features as a manifold. This design aims to explicitly disentangle domain-invariant features and domain-specific features while gaining individual control of both. The image translation process is formulated as a stylisation task, where the input is "stylised" (translated) into diverse target-domain images based on style codes sampled from the learnt manifold. We test MDGAN for multi-modal medical image translation, where we create two domain-specific manifold clusters on the manifold to translate segmentation maps into pseudo-CT and pseudo-MR images, respectively. We show that by traversing a path across the MR manifold cluster, the target output can be manipulated while still retaining the shape information from the input.
Fabric Image Representation Encoding Networks for Large-scale 3D Medical Image Analysis
Jun 30, 2020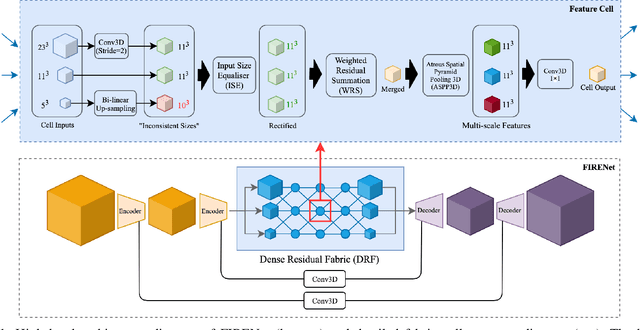
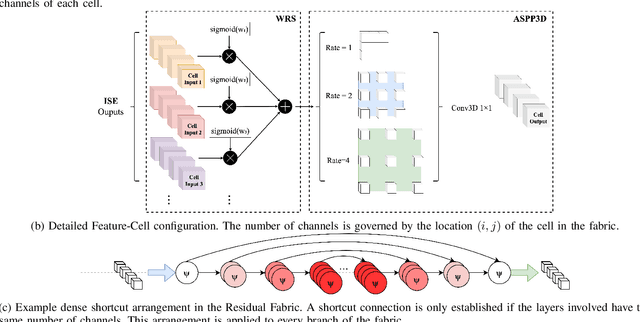
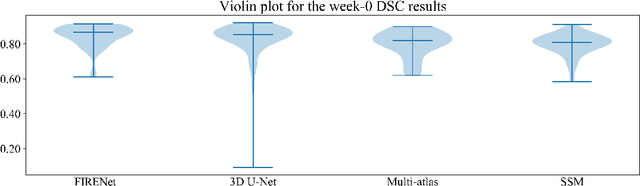
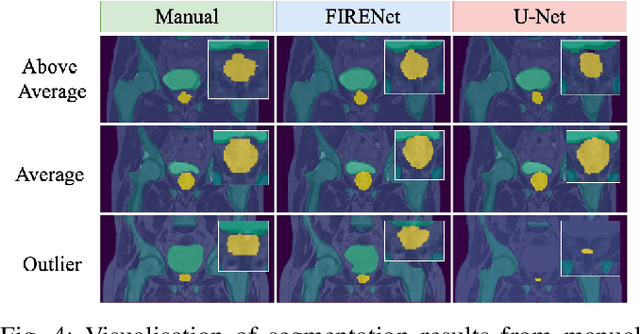
Abstract:Deep neural networks are parameterised by weights that encode feature representations, whose performance is dictated through generalisation by using large-scale feature-rich datasets. The lack of large-scale labelled 3D medical imaging datasets restrict constructing such generalised networks. In this work, a novel 3D segmentation network, Fabric Image Representation Networks (FIRENet), is proposed to extract and encode generalisable feature representations from multiple medical image datasets in a large-scale manner. FIRENet learns image specific feature representations by way of 3D fabric network architecture that contains exponential number of sub-architectures to handle various protocols and coverage of anatomical regions and structures. The fabric network uses Atrous Spatial Pyramid Pooling (ASPP) extended to 3D to extract local and image-level features at a fine selection of scales. The fabric is constructed with weighted edges allowing the learnt features to dynamically adapt to the training data at an architecture level. Conditional padding modules, which are integrated into the network to reinsert voxels discarded by feature pooling, allow the network to inherently process different-size images at their original resolutions. FIRENet was trained for feature learning via automated semantic segmentation of pelvic structures and obtained a state-of-the-art median DSC score of 0.867. FIRENet was also simultaneously trained on MR (Magnatic Resonance) images acquired from 3D examinations of musculoskeletal elements in the (hip, knee, shoulder) joints and a public OAI knee dataset to perform automated segmentation of bone across anatomy. Transfer learning was used to show that the features learnt through the pelvic segmentation helped achieve improved mean DSC scores of 0.962, 0.963, 0.945 and 0.986 for automated segmentation of bone across datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge