Stuart Crozier
Automated anomaly-aware 3D segmentation of bones and cartilages in knee MR images from the Osteoarthritis Initiative
Dec 01, 2022
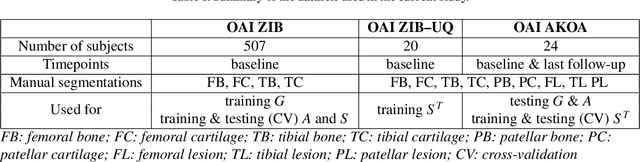
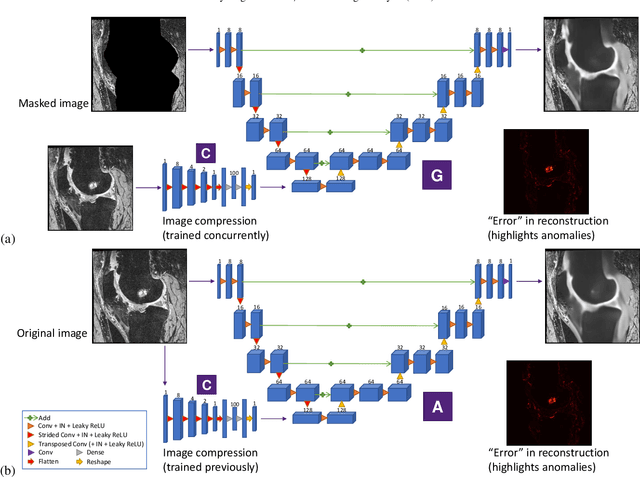
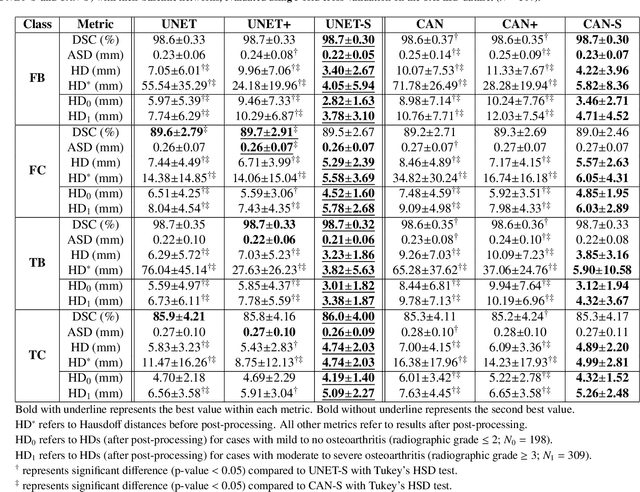
Abstract:In medical image analysis, automated segmentation of multi-component anatomical structures, which often have a spectrum of potential anomalies and pathologies, is a challenging task. In this work, we develop a multi-step approach using U-Net-based neural networks to initially detect anomalies (bone marrow lesions, bone cysts) in the distal femur, proximal tibia and patella from 3D magnetic resonance (MR) images of the knee in individuals with varying grades of osteoarthritis. Subsequently, the extracted data are used for downstream tasks involving semantic segmentation of individual bone and cartilage volumes as well as bone anomalies. For anomaly detection, the U-Net-based models were developed to reconstruct the bone profiles of the femur and tibia in images via inpainting so anomalous bone regions could be replaced with close to normal appearances. The reconstruction error was used to detect bone anomalies. A second anomaly-aware network, which was compared to anomaly-na\"ive segmentation networks, was used to provide a final automated segmentation of the femoral, tibial and patellar bones and cartilages from the knee MR images containing a spectrum of bone anomalies. The anomaly-aware segmentation approach provided up to 58% reduction in Hausdorff distances for bone segmentations compared to the results from the anomaly-na\"ive segmentation networks. In addition, the anomaly-aware networks were able to detect bone lesions in the MR images with greater sensitivity and specificity (area under the receiver operating characteristic curve [AUC] up to 0.896) compared to the anomaly-na\"ive segmentation networks (AUC up to 0.874).
Structure Guided Manifolds for Discovery of Disease Characteristics
Sep 24, 2022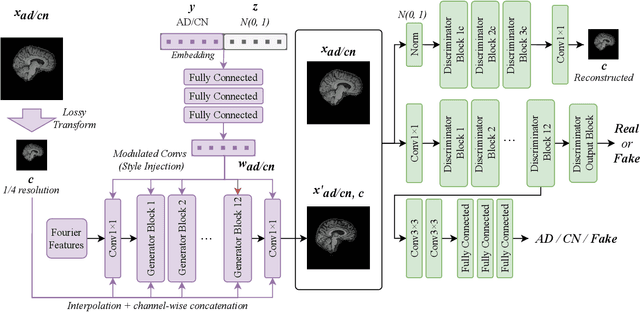
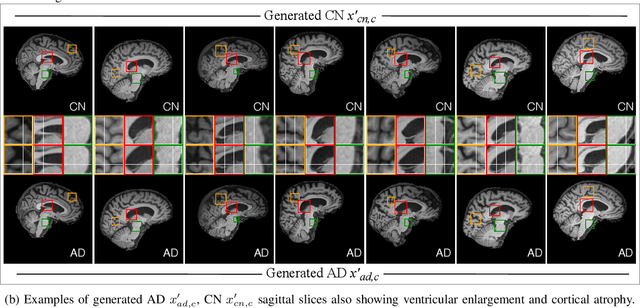
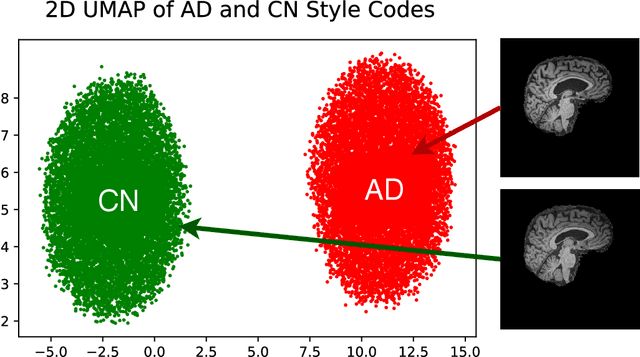

Abstract:In medical image analysis, the subtle visual characteristics of many diseases are challenging to discern, particularly due to the lack of paired data. For example, in mild Alzheimer's Disease (AD), brain tissue atrophy can be difficult to observe from pure imaging data, especially without paired AD and Cognitively Normal ( CN ) data for comparison. This work presents Disease Discovery GAN ( DiDiGAN), a weakly-supervised style-based framework for discovering and visualising subtle disease features. DiDiGAN learns a disease manifold of AD and CN visual characteristics, and the style codes sampled from this manifold are imposed onto an anatomical structural "blueprint" to synthesise paired AD and CN magnetic resonance images (MRIs). To suppress non-disease-related variations between the generated AD and CN pairs, DiDiGAN leverages a structural constraint with cycle consistency and anti-aliasing to enforce anatomical correspondence. When tested on the Alzheimer's Disease Neuroimaging Initiative ( ADNI) dataset, DiDiGAN showed key AD characteristics (reduced hippocampal volume, ventricular enlargement, and atrophy of cortical structures) through synthesising paired AD and CN scans. The qualitative results were backed up by automated brain volume analysis, where systematic pair-wise reductions in brain tissue structures were also measured
BFRnet: A deep learning-based MR background field removal method for QSM of the brain containing significant pathological susceptibility sources
Apr 06, 2022



Abstract:Introduction: Background field removal (BFR) is a critical step required for successful quantitative susceptibility mapping (QSM). However, eliminating the background field in brains containing significant susceptibility sources, such as intracranial hemorrhages, is challenging due to the relatively large scale of the field induced by these pathological susceptibility sources. Method: This study proposes a new deep learning-based method, BFRnet, to remove background field in healthy and hemorrhagic subjects. The network is built with the dual-frequency octave convolutions on the U-net architecture, trained with synthetic field maps containing significant susceptibility sources. The BFRnet method is compared with three conventional BFR methods and one previous deep learning method using simulated and in vivo brains from 4 healthy and 2 hemorrhagic subjects. Robustness against acquisition field-of-view (FOV) orientation and brain masking are also investigated. Results: For both simulation and in vivo experiments, BFRnet led to the best visually appealing results in the local field and QSM results with the minimum contrast loss and the most accurate hemorrhage susceptibility measurements among all five methods. In addition, BFRnet produced the most consistent local field and susceptibility maps between different sizes of brain masks, while conventional methods depend drastically on precise brain extraction and further brain edge erosions. It is also observed that BFRnet performed the best among all BFR methods for acquisition FOVs oblique to the main magnetic field. Conclusion: The proposed BFRnet improved the accuracy of local field reconstruction in the hemorrhagic subjects compared with conventional BFR algorithms. The BFRnet method was effective for acquisitions of titled orientations and retained whole brains without edge erosion as often required by traditional BFR methods.
Automated volumetric and statistical shape assessment of cam-type morphology of the femoral head-neck region from 3D magnetic resonance images
Dec 06, 2021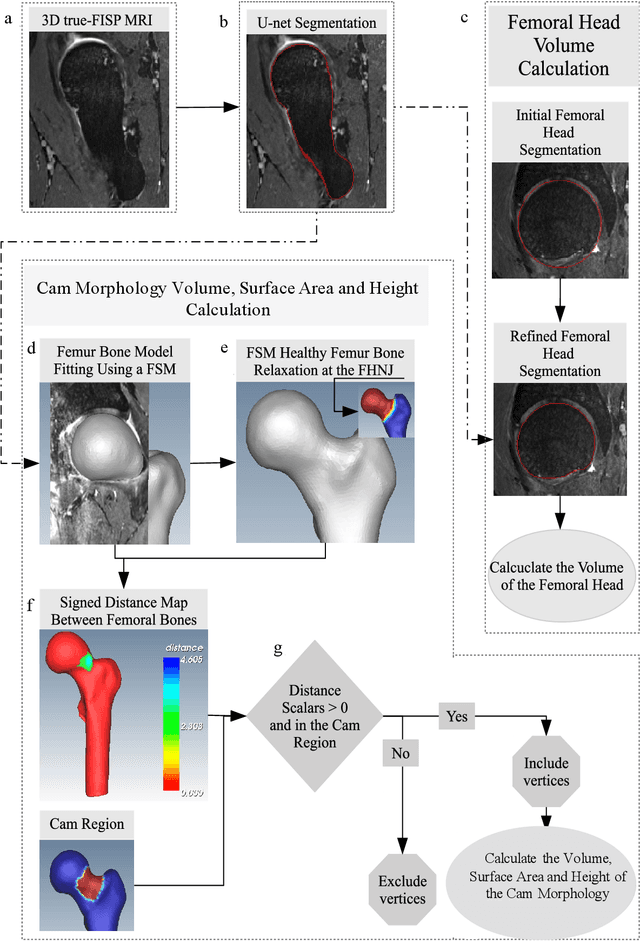
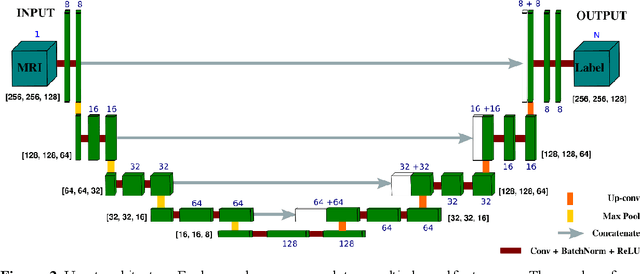

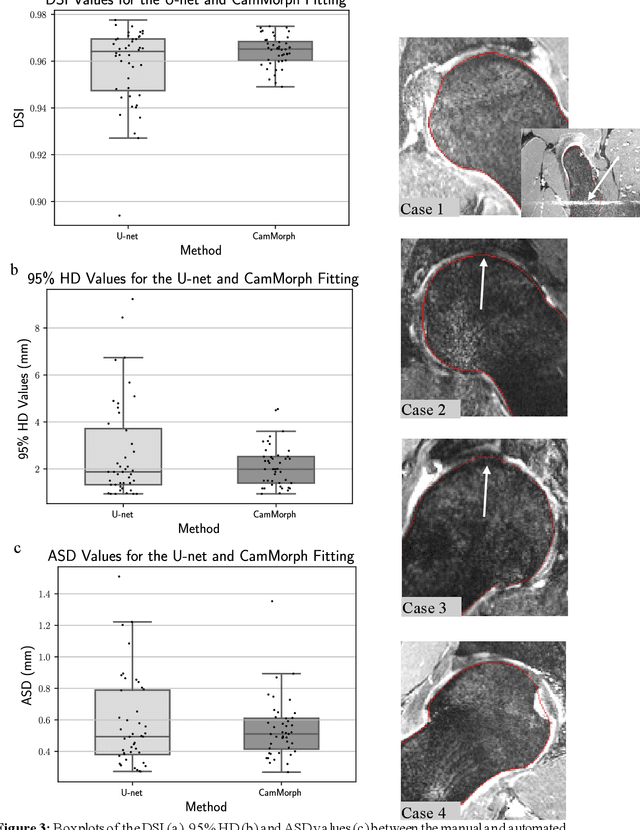
Abstract:Femoroacetabular impingement (FAI) cam morphology is routinely assessed using two-dimensional alpha angles which do not provide specific data on cam size characteristics. The purpose of this study is to implement a novel, automated three-dimensional (3D) pipeline, CamMorph, for segmentation and measurement of cam volume, surface area and height from magnetic resonance (MR) images in patients with FAI. The CamMorph pipeline involves two processes: i) proximal femur segmentation using an approach integrating 3D U-net with focused shape modelling (FSM); ii) use of patient-specific anatomical information from 3D FSM to simulate healthy femoral bone models and pathological region constraints to identify cam bone mass. Agreement between manual and automated segmentation of the proximal femur was evaluated with the Dice similarity index (DSI) and surface distance measures. Independent t-tests or Mann-Whitney U rank tests were used to compare the femoral head volume, cam volume, surface area and height data between female and male patients with FAI. There was a mean DSI value of 0.964 between manual and automated segmentation of proximal femur volume. Compared to female FAI patients, male patients had a significantly larger mean femoral head volume (66.12cm3 v 46.02cm3, p<0.001). Compared to female FAI patients, male patients had a significantly larger mean cam volume (1136.87mm3 v 337.86mm3, p<0.001), surface area (657.36mm2 v 306.93mm2 , p<0.001), maximum-height (3.89mm v 2.23mm, p<0.001) and average-height (1.94mm v 1.00mm, p<0.001). Automated analyses of 3D MR images from patients with FAI using the CamMorph pipeline showed that, in comparison with female patients, male patients had significantly greater cam volume, surface area and height.
Instant tissue field and magnetic susceptibility mapping from MR raw phase using Laplacian enabled deep neural networks
Nov 16, 2021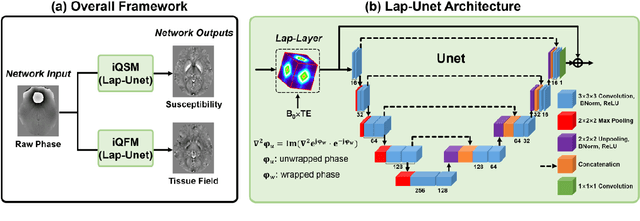

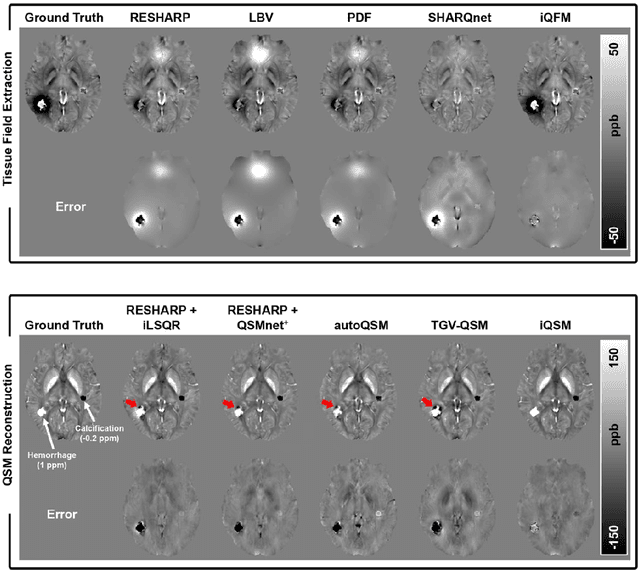

Abstract:Quantitative susceptibility mapping (QSM) is a valuable MRI post-processing technique that quantifies the magnetic susceptibility of body tissue from phase data. However, the traditional QSM reconstruction pipeline involves multiple non-trivial steps, including phase unwrapping, background field removal, and dipole inversion. These intermediate steps not only increase the reconstruction time but amplify noise and errors. This study develops a large-stencil Laplacian preprocessed deep learning-based neural network for near instant quantitative field and susceptibility mapping (i.e., iQFM and iQSM) from raw MR phase data. The proposed iQFM and iQSM methods were compared with established reconstruction pipelines on simulated and in vivo datasets. In addition, experiments on patients with intracranial hemorrhage and multiple sclerosis were also performed to test the generalization of the novel neural networks. The proposed iQFM and iQSM methods yielded comparable results to multi-step methods in healthy subjects while dramatically improving reconstruction accuracies on intracranial hemorrhages with large susceptibilities. The reconstruction time was also substantially shortened from minutes using multi-step methods to only 30 milliseconds using the trained iQFM and iQSM neural networks.
CAN3D: Fast 3D Medical Image Segmentation via Compact Context Aggregation
Sep 22, 2021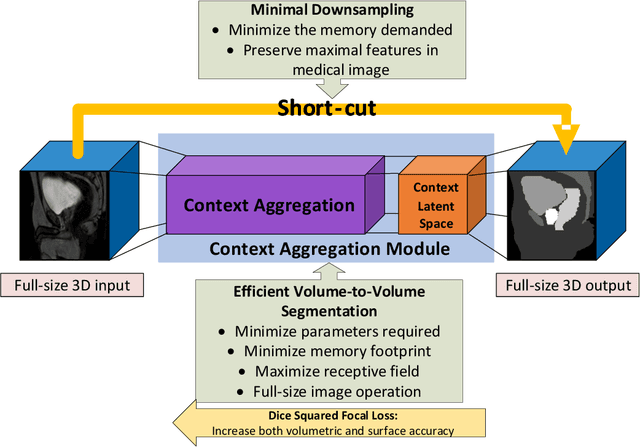

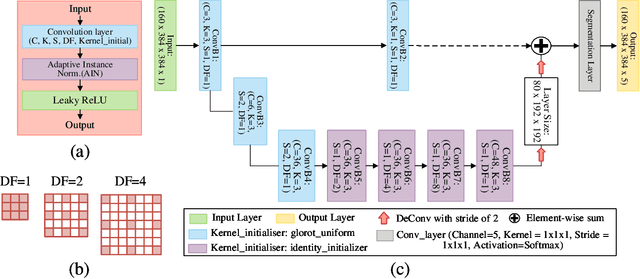
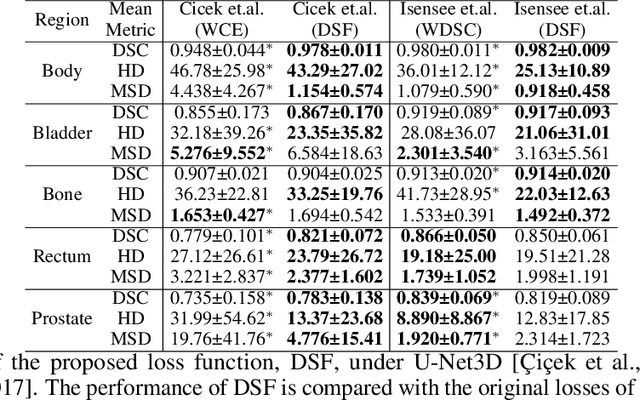
Abstract:Direct automatic segmentation of objects from 3D medical imaging, such as magnetic resonance (MR) imaging, is challenging as it often involves accurately identifying a number of individual objects with complex geometries within a large volume under investigation. To address these challenges, most deep learning approaches typically enhance their learning capability by substantially increasing the complexity or the number of trainable parameters within their models. Consequently, these models generally require long inference time on standard workstations operating clinical MR systems and are restricted to high-performance computing hardware due to their large memory requirement. Further, to fit 3D dataset through these large models using limited computer memory, trade-off techniques such as patch-wise training are often used which sacrifice the fine-scale geometric information from input images which could be clinically significant for diagnostic purposes. To address these challenges, we present a compact convolutional neural network with a shallow memory footprint to efficiently reduce the number of model parameters required for state-of-art performance. This is critical for practical employment as most clinical environments only have low-end hardware with limited computing power and memory. The proposed network can maintain data integrity by directly processing large full-size 3D input volumes with no patches required and significantly reduces the computational time required for both training and inference. We also propose a novel loss function with extra shape constraint to improve the accuracy for imbalanced classes in 3D MR images.
Bespoke Fractal Sampling Patterns for Discrete Fourier Space via the Kaleidoscope Transform
Aug 02, 2021



Abstract:Sampling strategies are important for sparse imaging methodologies, especially those employing the discrete Fourier transform (DFT). Chaotic sensing is one such methodology that employs deterministic, fractal sampling in conjunction with finite, iterative reconstruction schemes to form an image from limited samples. Using a sampling pattern constructed entirely from periodic lines in DFT space, chaotic sensing was found to outperform traditional compressed sensing for magnetic resonance imaging; however, only one such sampling pattern was presented and the reason for its fractal nature was not proven. Through the introduction of a novel image transform known as the kaleidoscope transform, which formalises and extends upon the concept of downsampling and concatenating an image with itself, this paper: (1) demonstrates a fundamental relationship between multiplication in modular arithmetic and downsampling; (2) provides a rigorous mathematical explanation for the fractal nature of the sampling pattern in the DFT; and (3) leverages this understanding to develop a collection of novel fractal sampling patterns for the 2D DFT with customisable properties. The ability to design tailor-made fractal sampling patterns expands the utility of the DFT in chaotic imaging and may form the basis for a bespoke chaotic sensing methodology, in which the fractal sampling matches the imaging task for improved reconstruction.
Deep Simultaneous Optimisation of Sampling and Reconstruction for Multi-contrast MRI
Mar 31, 2021



Abstract:MRI images of the same subject in different contrasts contain shared information, such as the anatomical structure. Utilizing the redundant information amongst the contrasts to sub-sample and faithfully reconstruct multi-contrast images could greatly accelerate the imaging speed, improve image quality and shorten scanning protocols. We propose an algorithm that generates the optimised sampling pattern and reconstruction scheme of one contrast (e.g. T2-weighted image) when images with different contrast (e.g. T1-weighted image) have been acquired. The proposed algorithm achieves increased PSNR and SSIM with the resulting optimal sampling pattern compared to other acquisition patterns and single contrast methods.
Accelerating Quantitative Susceptibility Mapping using Compressed Sensing and Deep Neural Network
Mar 17, 2021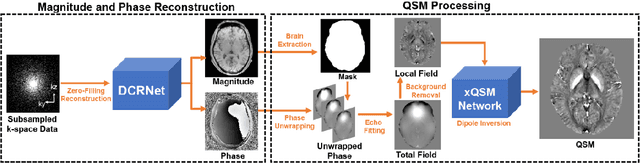
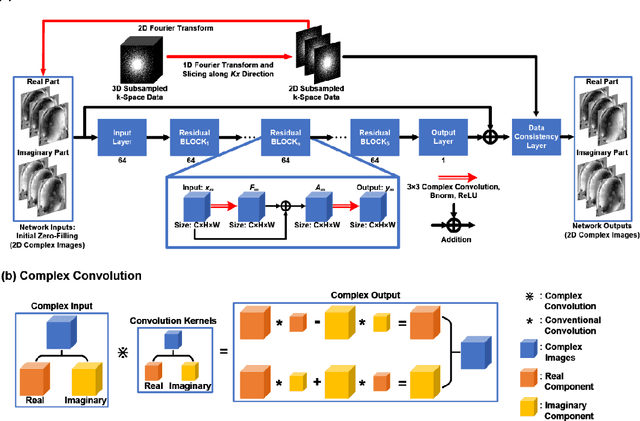
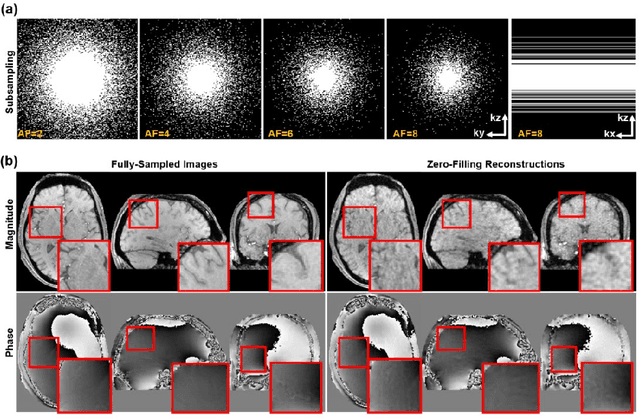
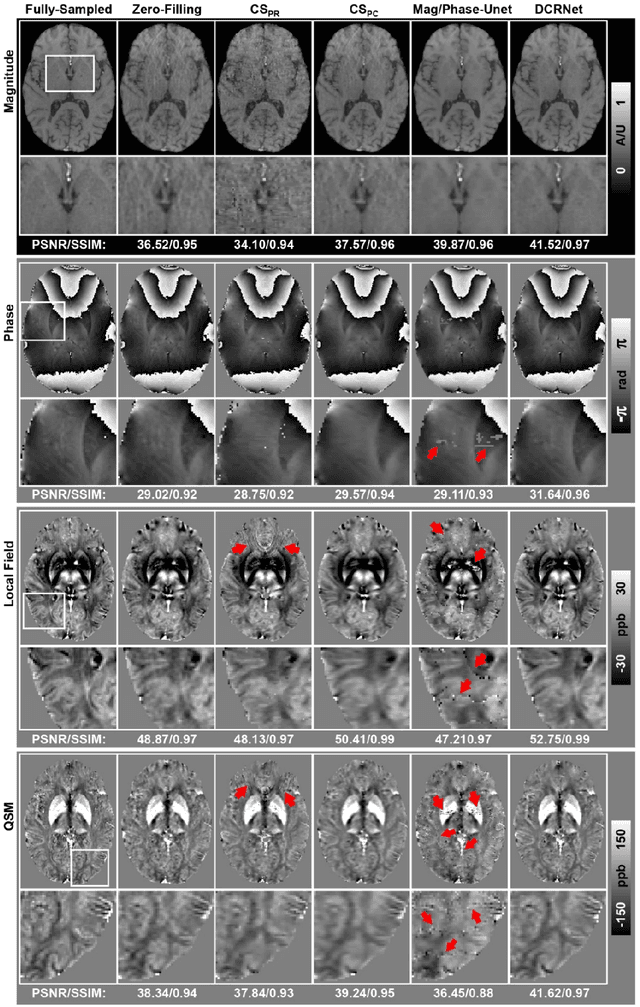
Abstract:Quantitative susceptibility mapping (QSM) is an MRI phase-based post-processing method that quantifies tissue magnetic susceptibility distributions. However, QSM acquisitions are relatively slow, even with parallel imaging. Incoherent undersampling and compressed sensing reconstruction techniques have been used to accelerate traditional magnitude-based MRI acquisitions; however, most do not recover the full phase signal due to its non-convex nature. In this study, a learning-based Deep Complex Residual Network (DCRNet) is proposed to recover both the magnitude and phase images from incoherently undersampled data, enabling high acceleration of QSM acquisition. Magnitude, phase, and QSM results from DCRNet were compared with two iterative and one deep learning methods on retrospectively undersampled acquisitions from six healthy volunteers, one intracranial hemorrhage and one multiple sclerosis patients, as well as one prospectively undersampled healthy subject using a 7T scanner. Peak signal to noise ratio (PSNR), structural similarity (SSIM) and region-of-interest susceptibility measurements are reported for numerical comparisons. The proposed DCRNet method substantially reduced artifacts and blurring compared to the other methods and resulted in the highest PSNR and SSIM on the magnitude, phase, local field, and susceptibility maps. It led to 4.0% to 8.8% accuracy improvements in deep grey matter susceptibility than some existing methods, when the acquisition was accelerated four times. The proposed DCRNet also dramatically shortened the reconstruction time by nearly 10 thousand times for each scan, from around 80 hours using conventional approaches to only 30 seconds.
Manipulating Medical Image Translation with Manifold Disentanglement
Nov 27, 2020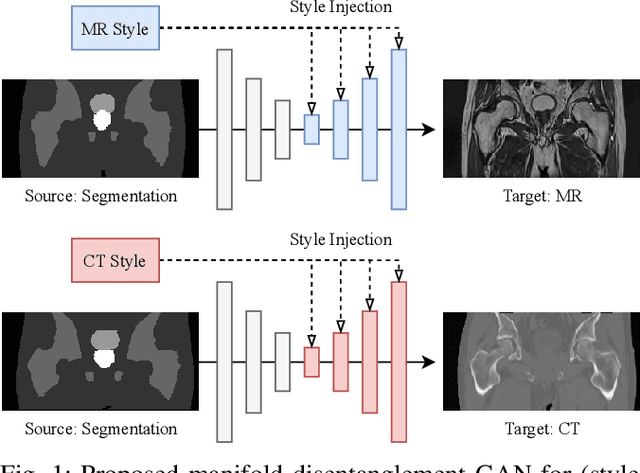
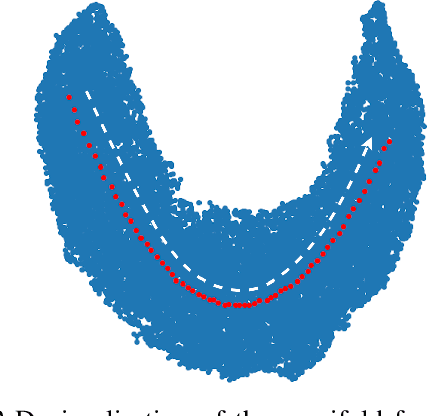

Abstract:Medical image translation (e.g. CT to MR) is a challenging task as it requires I) faithful translation of domain-invariant features (e.g. shape information of anatomical structures) and II) realistic synthesis of target-domain features (e.g. tissue appearance in MR). In this work, we propose Manifold Disentanglement Generative Adversarial Network (MDGAN), a novel image translation framework that explicitly models these two types of features. It employs a fully convolutional generator to model domain-invariant features, and it uses style codes to separately model target-domain features as a manifold. This design aims to explicitly disentangle domain-invariant features and domain-specific features while gaining individual control of both. The image translation process is formulated as a stylisation task, where the input is "stylised" (translated) into diverse target-domain images based on style codes sampled from the learnt manifold. We test MDGAN for multi-modal medical image translation, where we create two domain-specific manifold clusters on the manifold to translate segmentation maps into pseudo-CT and pseudo-MR images, respectively. We show that by traversing a path across the MR manifold cluster, the target output can be manipulated while still retaining the shape information from the input.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge