Jurgen Fripp
Gene-SGAN: a method for discovering disease subtypes with imaging and genetic signatures via multi-view weakly-supervised deep clustering
Jan 25, 2023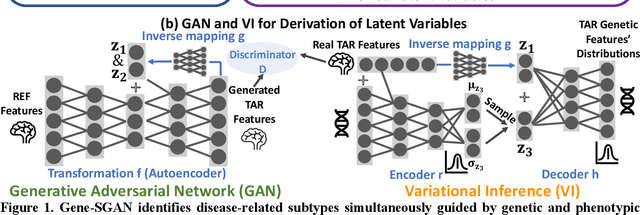
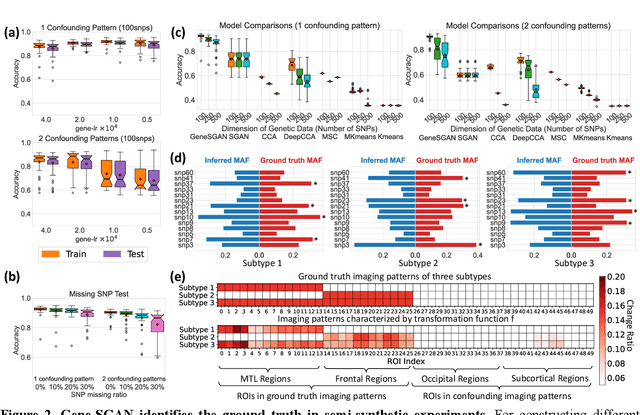
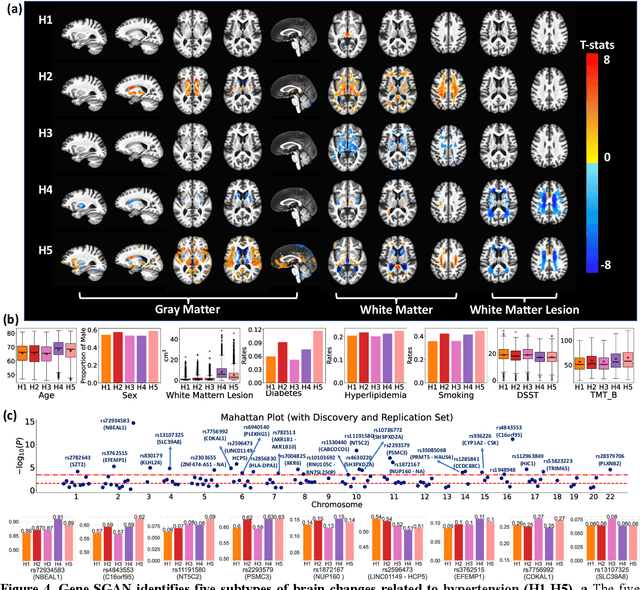
Abstract:Disease heterogeneity has been a critical challenge for precision diagnosis and treatment, especially in neurologic and neuropsychiatric diseases. Many diseases can display multiple distinct brain phenotypes across individuals, potentially reflecting disease subtypes that can be captured using MRI and machine learning methods. However, biological interpretability and treatment relevance are limited if the derived subtypes are not associated with genetic drivers or susceptibility factors. Herein, we describe Gene-SGAN - a multi-view, weakly-supervised deep clustering method - which dissects disease heterogeneity by jointly considering phenotypic and genetic data, thereby conferring genetic correlations to the disease subtypes and associated endophenotypic signatures. We first validate the generalizability, interpretability, and robustness of Gene-SGAN in semi-synthetic experiments. We then demonstrate its application to real multi-site datasets from 28,858 individuals, deriving subtypes of Alzheimer's disease and brain endophenotypes associated with hypertension, from MRI and SNP data. Derived brain phenotypes displayed significant differences in neuroanatomical patterns, genetic determinants, biological and clinical biomarkers, indicating potentially distinct underlying neuropathologic processes, genetic drivers, and susceptibility factors. Overall, Gene-SGAN is broadly applicable to disease subtyping and endophenotype discovery, and is herein tested on disease-related, genetically-driven neuroimaging phenotypes.
Automated anomaly-aware 3D segmentation of bones and cartilages in knee MR images from the Osteoarthritis Initiative
Dec 01, 2022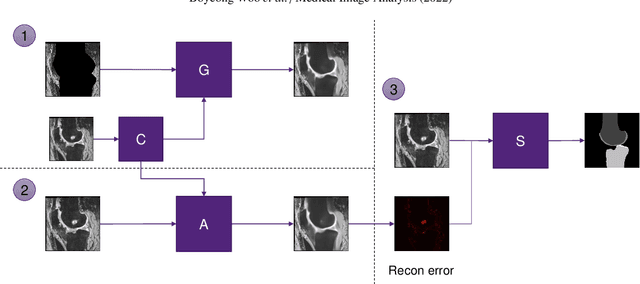
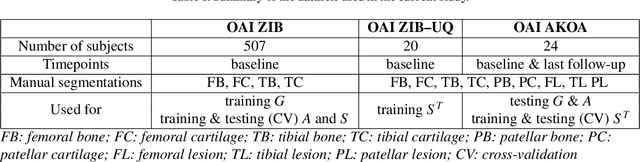
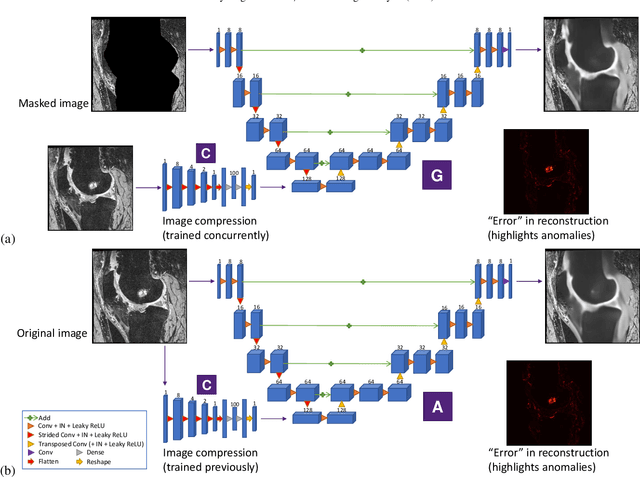
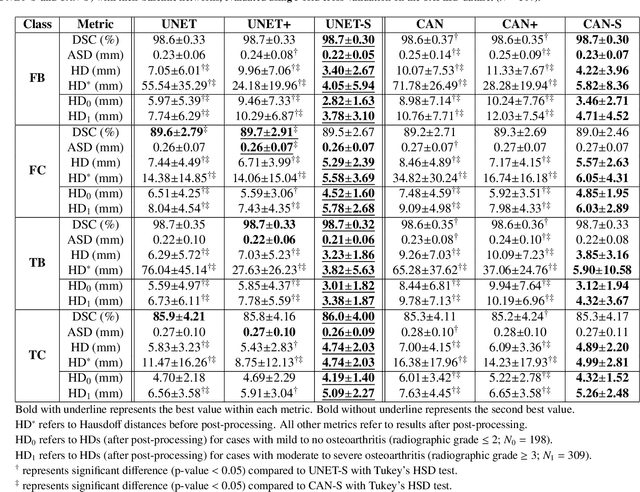
Abstract:In medical image analysis, automated segmentation of multi-component anatomical structures, which often have a spectrum of potential anomalies and pathologies, is a challenging task. In this work, we develop a multi-step approach using U-Net-based neural networks to initially detect anomalies (bone marrow lesions, bone cysts) in the distal femur, proximal tibia and patella from 3D magnetic resonance (MR) images of the knee in individuals with varying grades of osteoarthritis. Subsequently, the extracted data are used for downstream tasks involving semantic segmentation of individual bone and cartilage volumes as well as bone anomalies. For anomaly detection, the U-Net-based models were developed to reconstruct the bone profiles of the femur and tibia in images via inpainting so anomalous bone regions could be replaced with close to normal appearances. The reconstruction error was used to detect bone anomalies. A second anomaly-aware network, which was compared to anomaly-na\"ive segmentation networks, was used to provide a final automated segmentation of the femoral, tibial and patellar bones and cartilages from the knee MR images containing a spectrum of bone anomalies. The anomaly-aware segmentation approach provided up to 58% reduction in Hausdorff distances for bone segmentations compared to the results from the anomaly-na\"ive segmentation networks. In addition, the anomaly-aware networks were able to detect bone lesions in the MR images with greater sensitivity and specificity (area under the receiver operating characteristic curve [AUC] up to 0.896) compared to the anomaly-na\"ive segmentation networks (AUC up to 0.874).
CorticalFlow$^{++}$: Boosting Cortical Surface Reconstruction Accuracy, Regularity, and Interoperability
Jun 14, 2022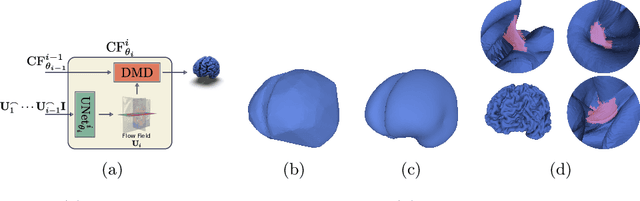
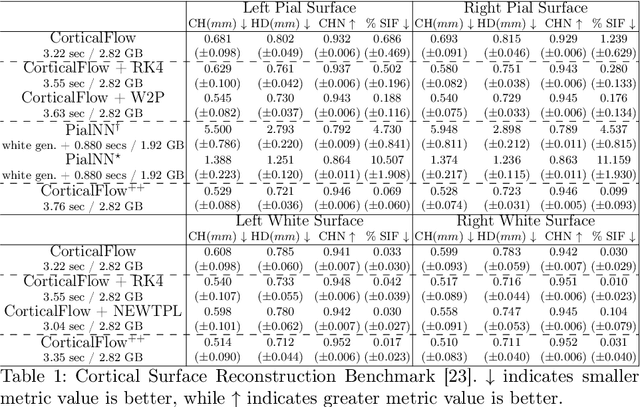
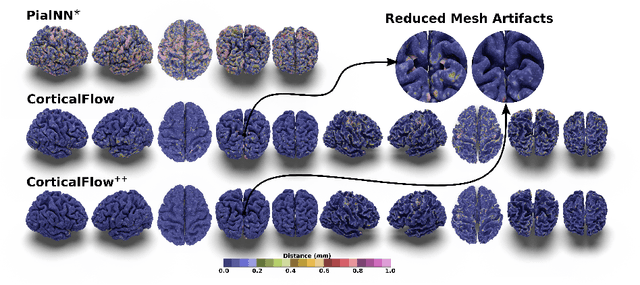
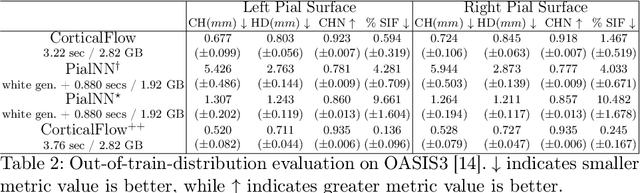
Abstract:The problem of Cortical Surface Reconstruction from magnetic resonance imaging has been traditionally addressed using lengthy pipelines of image processing techniques like FreeSurfer, CAT, or CIVET. These frameworks require very long runtimes deemed unfeasible for real-time applications and unpractical for large-scale studies. Recently, supervised deep learning approaches have been introduced to speed up this task cutting down the reconstruction time from hours to seconds. Using the state-of-the-art CorticalFlow model as a blueprint, this paper proposes three modifications to improve its accuracy and interoperability with existing surface analysis tools, while not sacrificing its fast inference time and low GPU memory consumption. First, we employ a more accurate ODE solver to reduce the diffeomorphic mapping approximation error. Second, we devise a routine to produce smoother template meshes avoiding mesh artifacts caused by sharp edges in CorticalFlow's convex-hull based template. Last, we recast pial surface prediction as the deformation of the predicted white surface leading to a one-to-one mapping between white and pial surface vertices. This mapping is essential to many existing surface analysis tools for cortical morphometry. We name the resulting method CorticalFlow$^{++}$. Using large-scale datasets, we demonstrate the proposed changes provide more geometric accuracy and surface regularity while keeping the reconstruction time and GPU memory requirements almost unchanged.
CorticalFlow: A Diffeomorphic Mesh Deformation Module for Cortical Surface Reconstruction
Jun 06, 2022
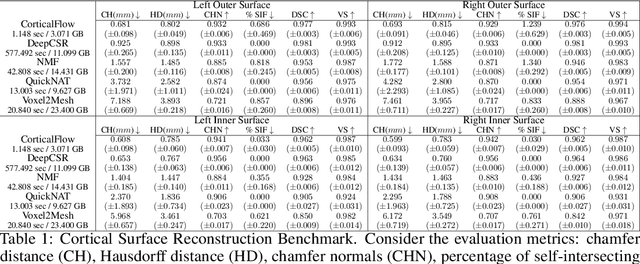
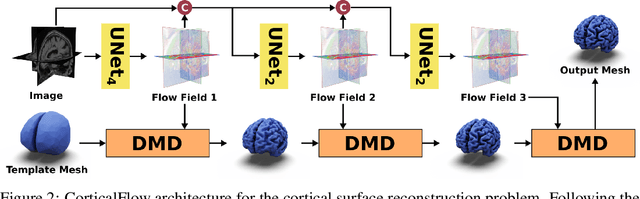
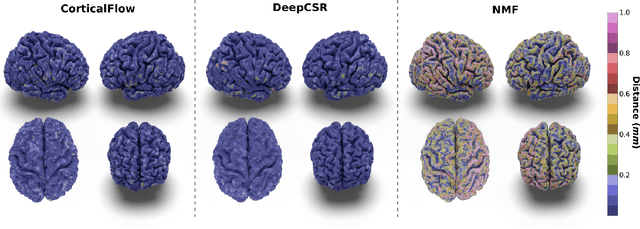
Abstract:In this paper we introduce CorticalFlow, a new geometric deep-learning model that, given a 3-dimensional image, learns to deform a reference template towards a targeted object. To conserve the template mesh's topological properties, we train our model over a set of diffeomorphic transformations. This new implementation of a flow Ordinary Differential Equation (ODE) framework benefits from a small GPU memory footprint, allowing the generation of surfaces with several hundred thousand vertices. To reduce topological errors introduced by its discrete resolution, we derive numeric conditions which improve the manifoldness of the predicted triangle mesh. To exhibit the utility of CorticalFlow, we demonstrate its performance for the challenging task of brain cortical surface reconstruction. In contrast to current state-of-the-art, CorticalFlow produces superior surfaces while reducing the computation time from nine and a half minutes to one second. More significantly, CorticalFlow enforces the generation of anatomically plausible surfaces; the absence of which has been a major impediment restricting the clinical relevance of such surface reconstruction methods.
Automated volumetric and statistical shape assessment of cam-type morphology of the femoral head-neck region from 3D magnetic resonance images
Dec 06, 2021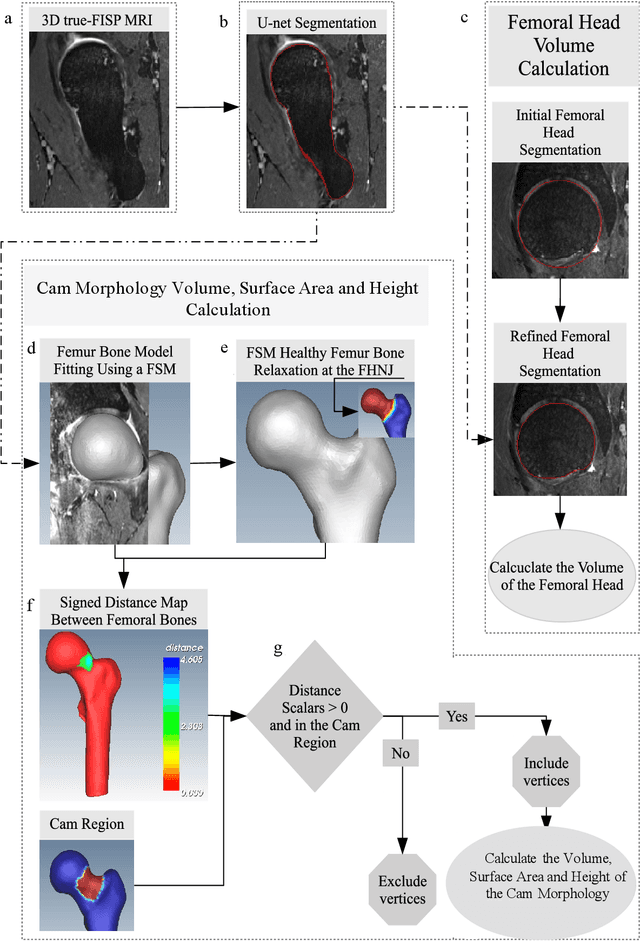
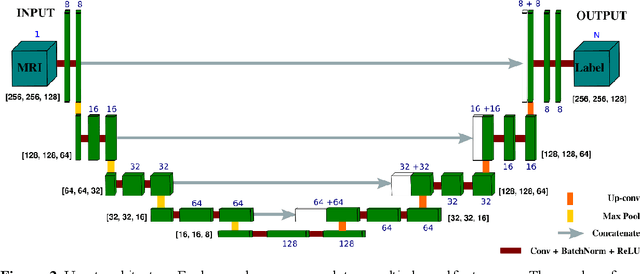

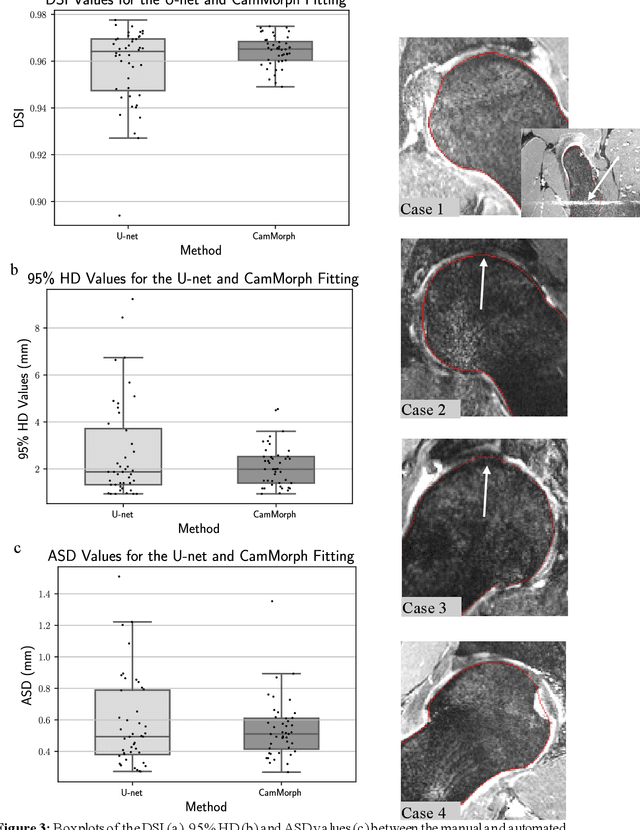
Abstract:Femoroacetabular impingement (FAI) cam morphology is routinely assessed using two-dimensional alpha angles which do not provide specific data on cam size characteristics. The purpose of this study is to implement a novel, automated three-dimensional (3D) pipeline, CamMorph, for segmentation and measurement of cam volume, surface area and height from magnetic resonance (MR) images in patients with FAI. The CamMorph pipeline involves two processes: i) proximal femur segmentation using an approach integrating 3D U-net with focused shape modelling (FSM); ii) use of patient-specific anatomical information from 3D FSM to simulate healthy femoral bone models and pathological region constraints to identify cam bone mass. Agreement between manual and automated segmentation of the proximal femur was evaluated with the Dice similarity index (DSI) and surface distance measures. Independent t-tests or Mann-Whitney U rank tests were used to compare the femoral head volume, cam volume, surface area and height data between female and male patients with FAI. There was a mean DSI value of 0.964 between manual and automated segmentation of proximal femur volume. Compared to female FAI patients, male patients had a significantly larger mean femoral head volume (66.12cm3 v 46.02cm3, p<0.001). Compared to female FAI patients, male patients had a significantly larger mean cam volume (1136.87mm3 v 337.86mm3, p<0.001), surface area (657.36mm2 v 306.93mm2 , p<0.001), maximum-height (3.89mm v 2.23mm, p<0.001) and average-height (1.94mm v 1.00mm, p<0.001). Automated analyses of 3D MR images from patients with FAI using the CamMorph pipeline showed that, in comparison with female patients, male patients had significantly greater cam volume, surface area and height.
DeepCSR: A 3D Deep Learning Approach for Cortical Surface Reconstruction
Oct 22, 2020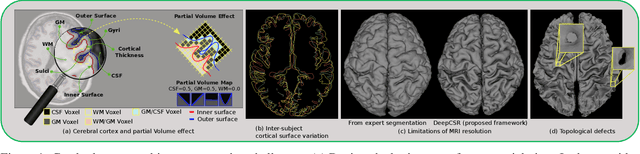
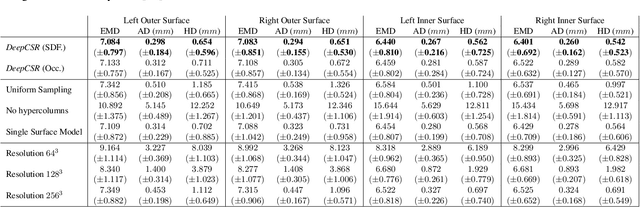

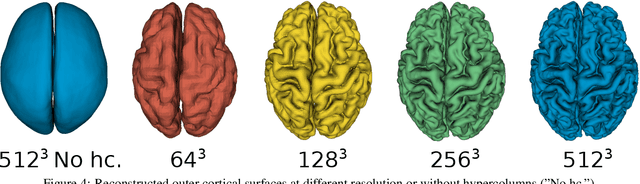
Abstract:The study of neurodegenerative diseases relies on the reconstruction and analysis of the brain cortex from magnetic resonance imaging (MRI). Traditional frameworks for this task like FreeSurfer demand lengthy runtimes, while its accelerated variant FastSurfer still relies on a voxel-wise segmentation which is limited by its resolution to capture narrow continuous objects as cortical surfaces. Having these limitations in mind, we propose DeepCSR, a 3D deep learning framework for cortical surface reconstruction from MRI. Towards this end, we train a neural network model with hypercolumn features to predict implicit surface representations for points in a brain template space. After training, the cortical surface at a desired level of detail is obtained by evaluating surface representations at specific coordinates, and subsequently applying a topology correction algorithm and an isosurface extraction method. Thanks to the continuous nature of this approach and the efficacy of its hypercolumn features scheme, DeepCSR efficiently reconstructs cortical surfaces at high resolution capturing fine details in the cortical folding. Moreover, DeepCSR is as accurate, more precise, and faster than the widely used FreeSurfer toolbox and its deep learning powered variant FastSurfer on reconstructing cortical surfaces from MRI which should facilitate large-scale medical studies and new healthcare applications.
Medical Image Harmonization Using Deep Learning Based Canonical Mapping: Toward Robust and Generalizable Learning in Imaging
Oct 11, 2020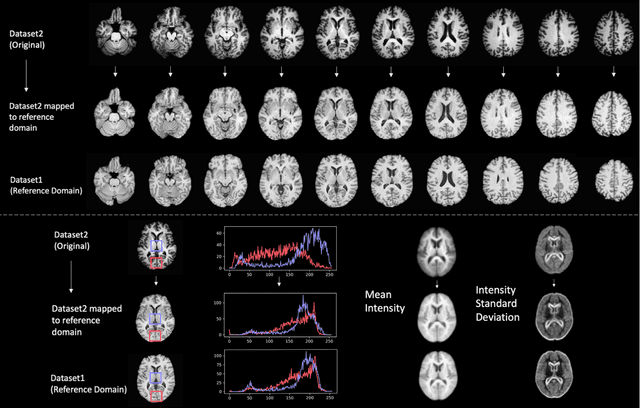
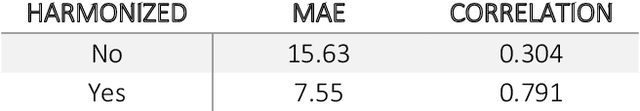
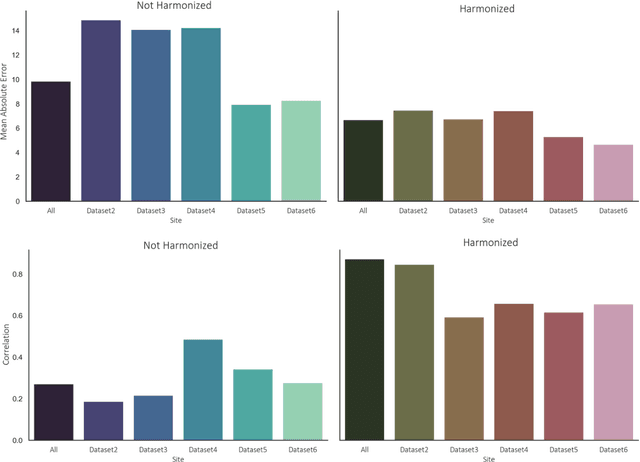
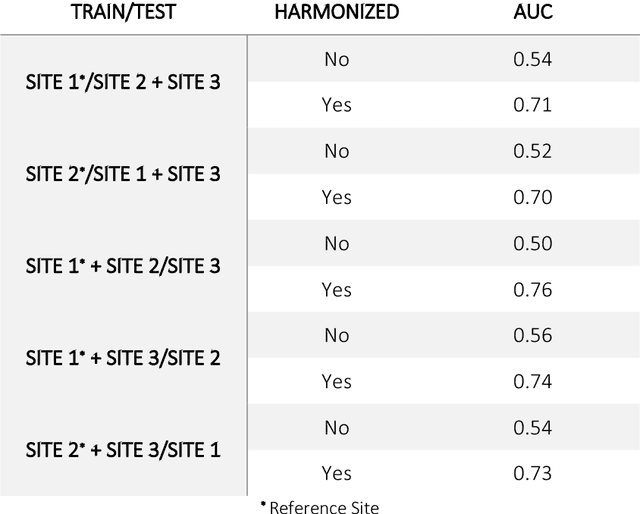
Abstract:Conventional and deep learning-based methods have shown great potential in the medical imaging domain, as means for deriving diagnostic, prognostic, and predictive biomarkers, and by contributing to precision medicine. However, these methods have yet to see widespread clinical adoption, in part due to limited generalization performance across various imaging devices, acquisition protocols, and patient populations. In this work, we propose a new paradigm in which data from a diverse range of acquisition conditions are "harmonized" to a common reference domain, where accurate model learning and prediction can take place. By learning an unsupervised image to image canonical mapping from diverse datasets to a reference domain using generative deep learning models, we aim to reduce confounding data variation while preserving semantic information, thereby rendering the learning task easier in the reference domain. We test this approach on two example problems, namely MRI-based brain age prediction and classification of schizophrenia, leveraging pooled cohorts of neuroimaging MRI data spanning 9 sites and 9701 subjects. Our results indicate a substantial improvement in these tasks in out-of-sample data, even when training is restricted to a single site.
Going deeper with brain morphometry using neural networks
Sep 07, 2020
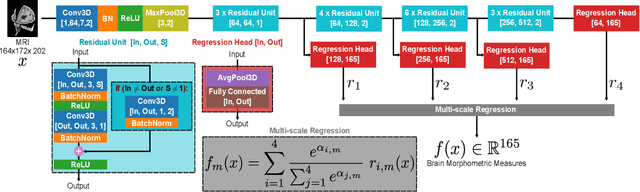
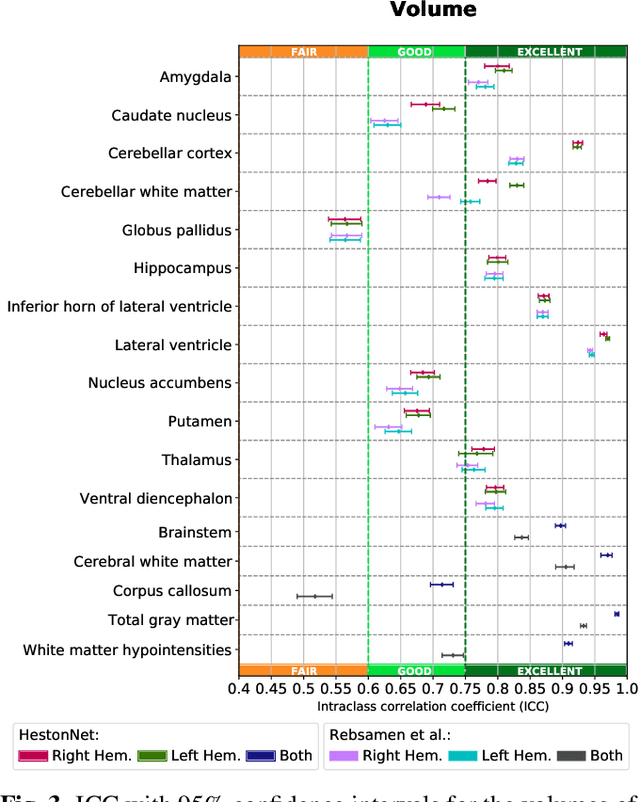
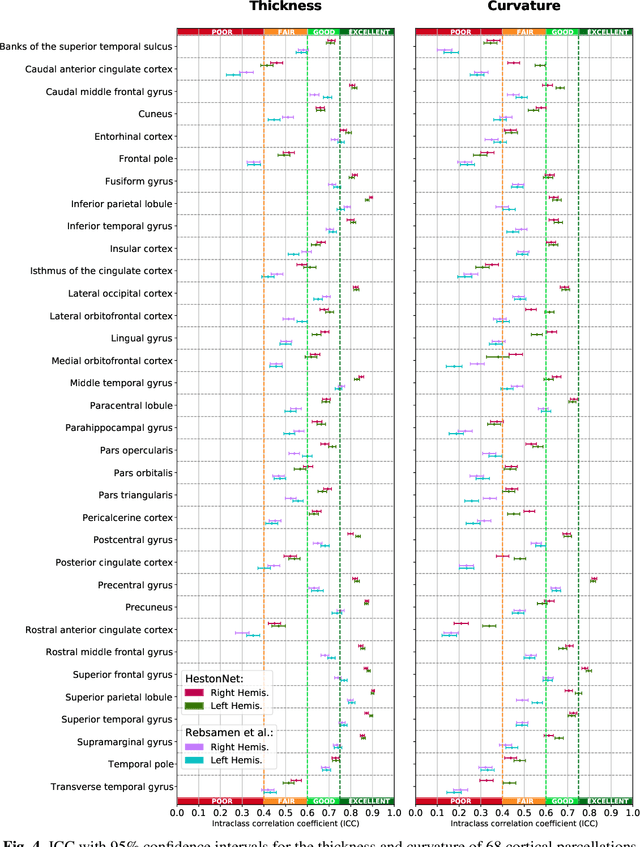
Abstract:Brain morphometry from magnetic resonance imaging (MRI) is a consolidated biomarker for many neurodegenerative diseases. Recent advances in this domain indicate that deep convolutional neural networks can infer morphometric measurements within a few seconds. Nevertheless, the accuracy of the devised model for insightful bio-markers (mean curvature and thickness) remains unsatisfactory. In this paper, we propose a more accurate and efficient neural network model for brain morphometry named HerstonNet. More specifically, we develop a 3D ResNet-based neural network to learn rich features directly from MRI, design a multi-scale regression scheme by predicting morphometric measures at feature maps of different resolutions, and leverage a robust optimization method to avoid poor quality minima and reduce the prediction variance. As a result, HerstonNet improves the existing approach by 24.30% in terms of intraclass correlation coefficient (agreement measure) to FreeSurfer silver-standards while maintaining a competitive run-time.
Fabric Image Representation Encoding Networks for Large-scale 3D Medical Image Analysis
Jun 30, 2020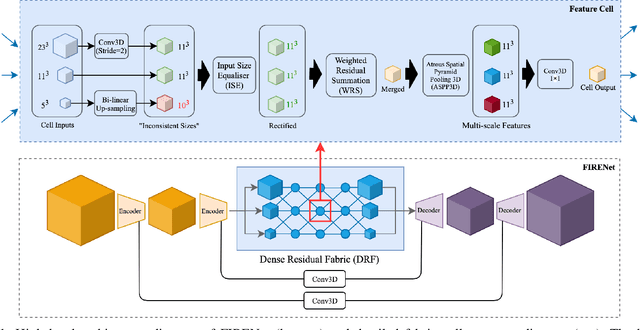
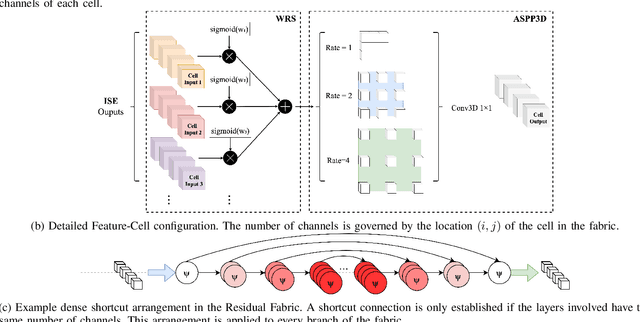
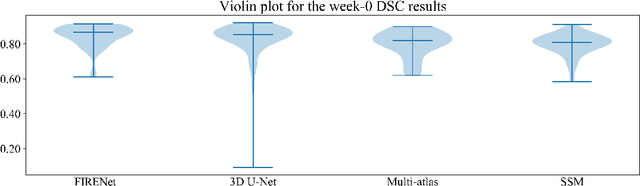
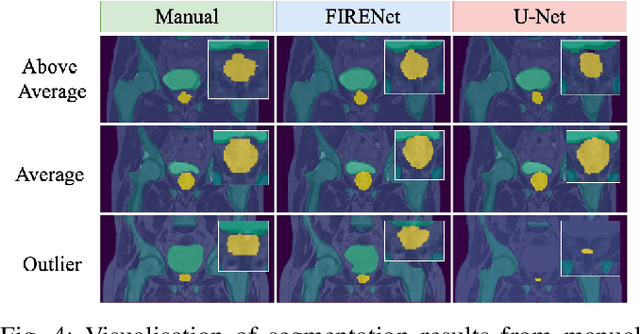
Abstract:Deep neural networks are parameterised by weights that encode feature representations, whose performance is dictated through generalisation by using large-scale feature-rich datasets. The lack of large-scale labelled 3D medical imaging datasets restrict constructing such generalised networks. In this work, a novel 3D segmentation network, Fabric Image Representation Networks (FIRENet), is proposed to extract and encode generalisable feature representations from multiple medical image datasets in a large-scale manner. FIRENet learns image specific feature representations by way of 3D fabric network architecture that contains exponential number of sub-architectures to handle various protocols and coverage of anatomical regions and structures. The fabric network uses Atrous Spatial Pyramid Pooling (ASPP) extended to 3D to extract local and image-level features at a fine selection of scales. The fabric is constructed with weighted edges allowing the learnt features to dynamically adapt to the training data at an architecture level. Conditional padding modules, which are integrated into the network to reinsert voxels discarded by feature pooling, allow the network to inherently process different-size images at their original resolutions. FIRENet was trained for feature learning via automated semantic segmentation of pelvic structures and obtained a state-of-the-art median DSC score of 0.867. FIRENet was also simultaneously trained on MR (Magnatic Resonance) images acquired from 3D examinations of musculoskeletal elements in the (hip, knee, shoulder) joints and a public OAI knee dataset to perform automated segmentation of bone across anatomy. Transfer learning was used to show that the features learnt through the pelvic segmentation helped achieve improved mean DSC scores of 0.962, 0.963, 0.945 and 0.986 for automated segmentation of bone across datasets.
3D Scanning System for Automatic High-Resolution Plant Phenotyping
Feb 26, 2017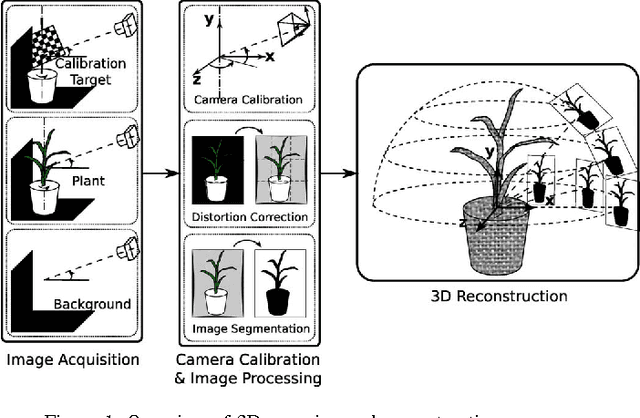
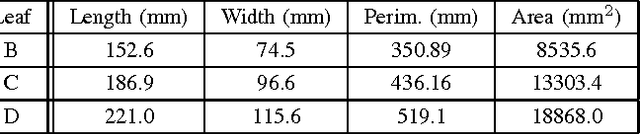
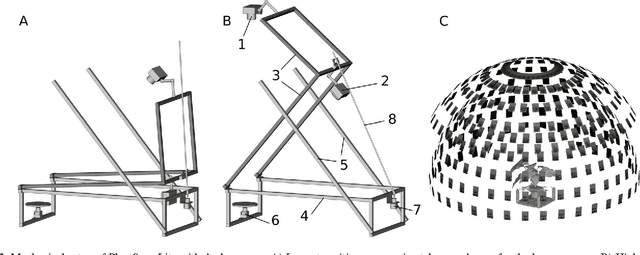
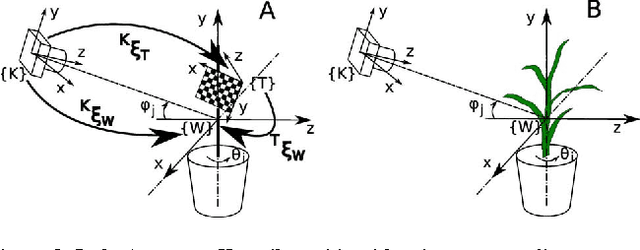
Abstract:Thin leaves, fine stems, self-occlusion, non-rigid and slowly changing structures make plants difficult for three-dimensional (3D) scanning and reconstruction -- two critical steps in automated visual phenotyping. Many current solutions such as laser scanning, structured light, and multiview stereo can struggle to acquire usable 3D models because of limitations in scanning resolution and calibration accuracy. In response, we have developed a fast, low-cost, 3D scanning platform to image plants on a rotating stage with two tilting DSLR cameras centred on the plant. This uses new methods of camera calibration and background removal to achieve high-accuracy 3D reconstruction. We assessed the system's accuracy using a 3D visual hull reconstruction algorithm applied on 2 plastic models of dicotyledonous plants, 2 sorghum plants and 2 wheat plants across different sets of tilt angles. Scan times ranged from 3 minutes (to capture 72 images using 2 tilt angles), to 30 minutes (to capture 360 images using 10 tilt angles). The leaf lengths, widths, areas and perimeters of the plastic models were measured manually and compared to measurements from the scanning system: results were within 3-4% of each other. The 3D reconstructions obtained with the scanning system show excellent geometric agreement with all six plant specimens, even plants with thin leaves and fine stems.
* 8 papes, DICTA 2016
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge