Pierrick Bourgeat
NC-Reg : Neural Cortical Maps for Rigid Registration
Jan 26, 2026Abstract:We introduce neural cortical maps, a continuous and compact neural representation for cortical feature maps, as an alternative to traditional discrete structures such as grids and meshes. It can learn from meshes of arbitrary size and provide learnt features at any resolution. Neural cortical maps enable efficient optimization on the sphere and achieve runtimes up to 30 times faster than classic barycentric interpolation (for the same number of iterations). As a proof of concept, we investigate rigid registration of cortical surfaces and propose NC-Reg, a novel iterative algorithm that involves the use of neural cortical feature maps, gradient descent optimization and a simulated annealing strategy. Through ablation studies and subject-to-template experiments, our method demonstrates sub-degree accuracy ($<1^\circ$ from the global optimum), and serves as a promising robust pre-alignment strategy, which is critical in clinical settings.
CorticalFlow$^{++}$: Boosting Cortical Surface Reconstruction Accuracy, Regularity, and Interoperability
Jun 14, 2022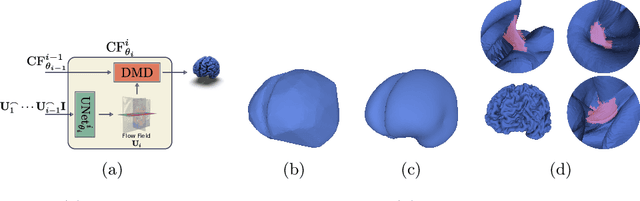
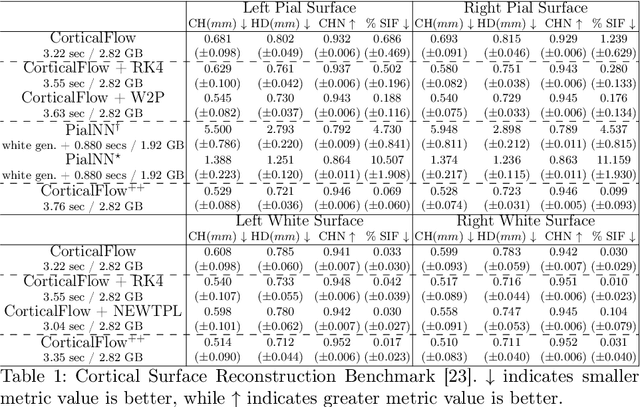
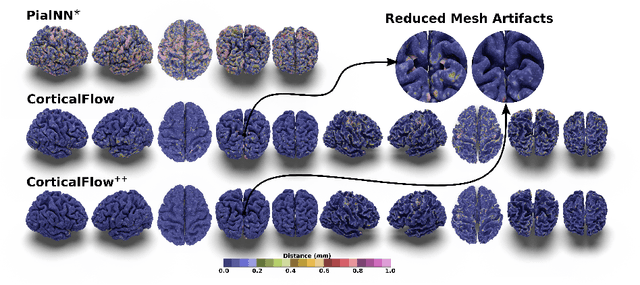
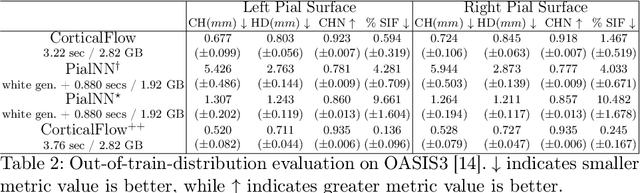
Abstract:The problem of Cortical Surface Reconstruction from magnetic resonance imaging has been traditionally addressed using lengthy pipelines of image processing techniques like FreeSurfer, CAT, or CIVET. These frameworks require very long runtimes deemed unfeasible for real-time applications and unpractical for large-scale studies. Recently, supervised deep learning approaches have been introduced to speed up this task cutting down the reconstruction time from hours to seconds. Using the state-of-the-art CorticalFlow model as a blueprint, this paper proposes three modifications to improve its accuracy and interoperability with existing surface analysis tools, while not sacrificing its fast inference time and low GPU memory consumption. First, we employ a more accurate ODE solver to reduce the diffeomorphic mapping approximation error. Second, we devise a routine to produce smoother template meshes avoiding mesh artifacts caused by sharp edges in CorticalFlow's convex-hull based template. Last, we recast pial surface prediction as the deformation of the predicted white surface leading to a one-to-one mapping between white and pial surface vertices. This mapping is essential to many existing surface analysis tools for cortical morphometry. We name the resulting method CorticalFlow$^{++}$. Using large-scale datasets, we demonstrate the proposed changes provide more geometric accuracy and surface regularity while keeping the reconstruction time and GPU memory requirements almost unchanged.
CorticalFlow: A Diffeomorphic Mesh Deformation Module for Cortical Surface Reconstruction
Jun 06, 2022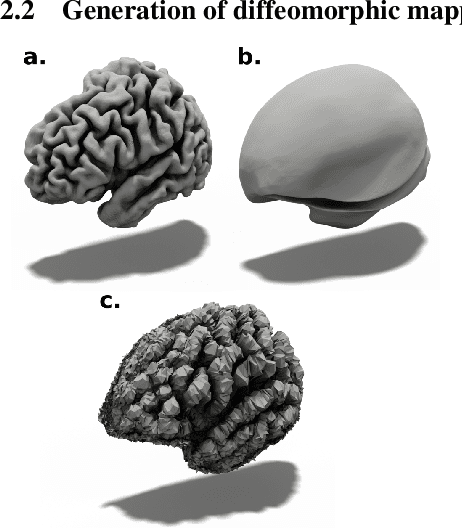
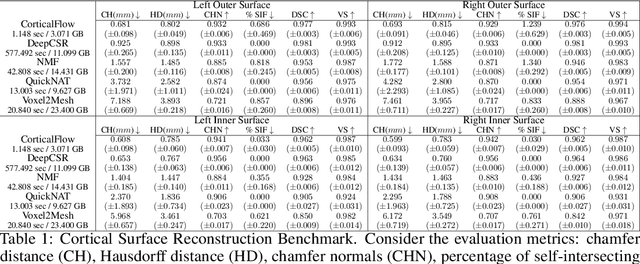
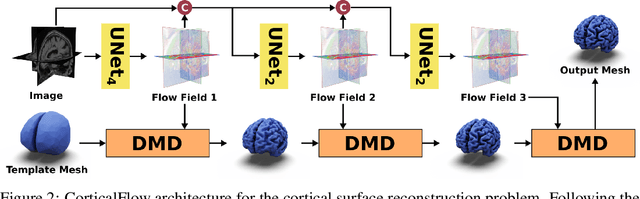
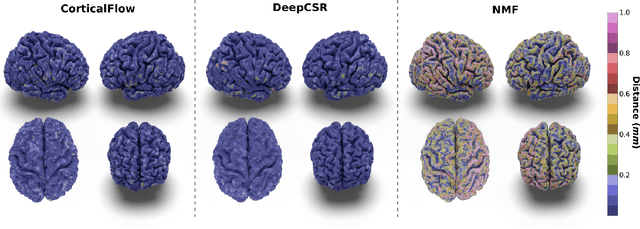
Abstract:In this paper we introduce CorticalFlow, a new geometric deep-learning model that, given a 3-dimensional image, learns to deform a reference template towards a targeted object. To conserve the template mesh's topological properties, we train our model over a set of diffeomorphic transformations. This new implementation of a flow Ordinary Differential Equation (ODE) framework benefits from a small GPU memory footprint, allowing the generation of surfaces with several hundred thousand vertices. To reduce topological errors introduced by its discrete resolution, we derive numeric conditions which improve the manifoldness of the predicted triangle mesh. To exhibit the utility of CorticalFlow, we demonstrate its performance for the challenging task of brain cortical surface reconstruction. In contrast to current state-of-the-art, CorticalFlow produces superior surfaces while reducing the computation time from nine and a half minutes to one second. More significantly, CorticalFlow enforces the generation of anatomically plausible surfaces; the absence of which has been a major impediment restricting the clinical relevance of such surface reconstruction methods.
DeepCSR: A 3D Deep Learning Approach for Cortical Surface Reconstruction
Oct 22, 2020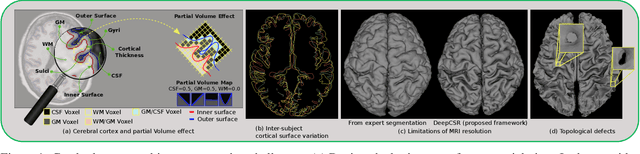
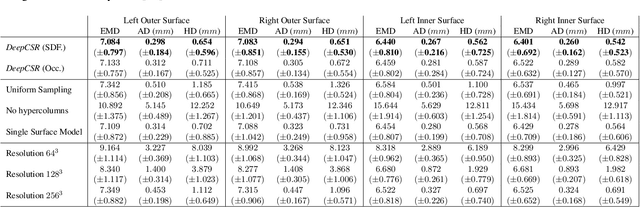

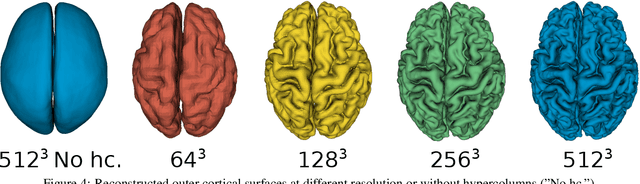
Abstract:The study of neurodegenerative diseases relies on the reconstruction and analysis of the brain cortex from magnetic resonance imaging (MRI). Traditional frameworks for this task like FreeSurfer demand lengthy runtimes, while its accelerated variant FastSurfer still relies on a voxel-wise segmentation which is limited by its resolution to capture narrow continuous objects as cortical surfaces. Having these limitations in mind, we propose DeepCSR, a 3D deep learning framework for cortical surface reconstruction from MRI. Towards this end, we train a neural network model with hypercolumn features to predict implicit surface representations for points in a brain template space. After training, the cortical surface at a desired level of detail is obtained by evaluating surface representations at specific coordinates, and subsequently applying a topology correction algorithm and an isosurface extraction method. Thanks to the continuous nature of this approach and the efficacy of its hypercolumn features scheme, DeepCSR efficiently reconstructs cortical surfaces at high resolution capturing fine details in the cortical folding. Moreover, DeepCSR is as accurate, more precise, and faster than the widely used FreeSurfer toolbox and its deep learning powered variant FastSurfer on reconstructing cortical surfaces from MRI which should facilitate large-scale medical studies and new healthcare applications.
Going deeper with brain morphometry using neural networks
Sep 07, 2020
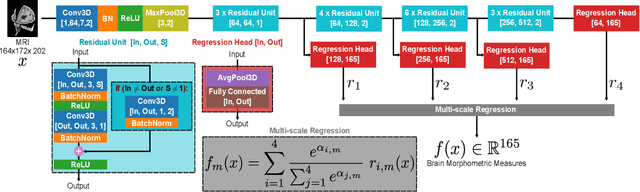
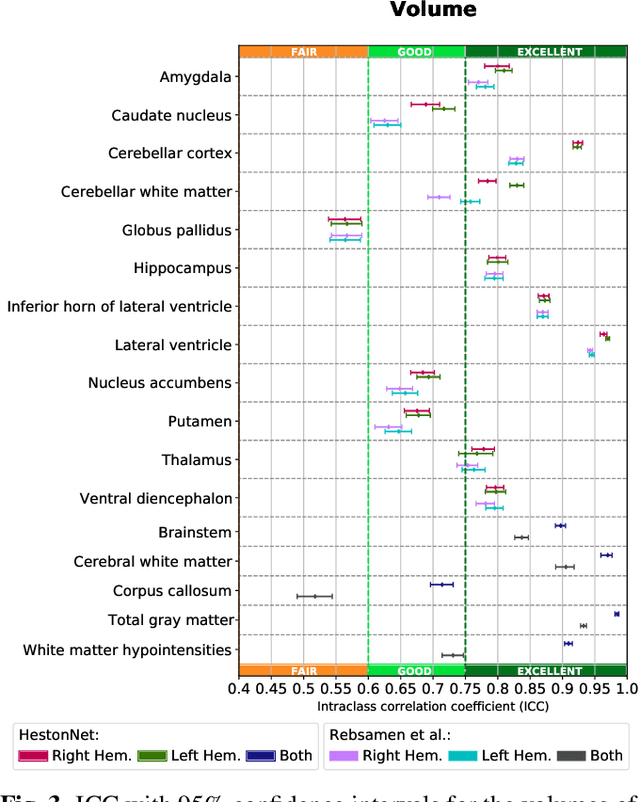
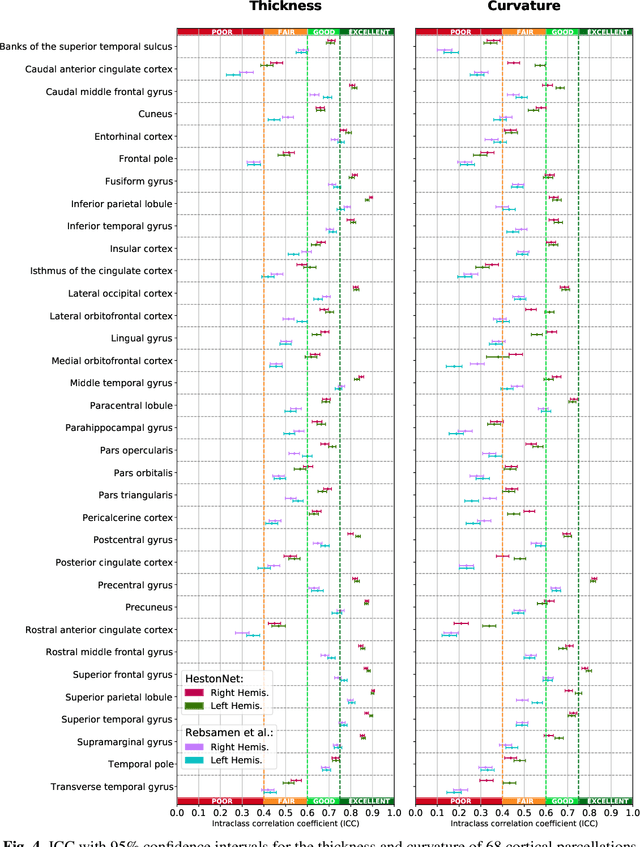
Abstract:Brain morphometry from magnetic resonance imaging (MRI) is a consolidated biomarker for many neurodegenerative diseases. Recent advances in this domain indicate that deep convolutional neural networks can infer morphometric measurements within a few seconds. Nevertheless, the accuracy of the devised model for insightful bio-markers (mean curvature and thickness) remains unsatisfactory. In this paper, we propose a more accurate and efficient neural network model for brain morphometry named HerstonNet. More specifically, we develop a 3D ResNet-based neural network to learn rich features directly from MRI, design a multi-scale regression scheme by predicting morphometric measures at feature maps of different resolutions, and leverage a robust optimization method to avoid poor quality minima and reduce the prediction variance. As a result, HerstonNet improves the existing approach by 24.30% in terms of intraclass correlation coefficient (agreement measure) to FreeSurfer silver-standards while maintaining a competitive run-time.
Enforcing temporal consistency in Deep Learning segmentation of brain MR images
Jun 13, 2019



Abstract:Longitudinal analysis has great potential to reveal developmental trajectories and monitor disease progression in medical imaging. This process relies on consistent and robust joint 4D segmentation. Traditional techniques are dependent on the similarity of images over time and the use of subject-specific priors to reduce random variation and improve the robustness and sensitivity of the overall longitudinal analysis. This is however slow and computationally intensive as subject-specific templates need to be rebuilt every time. The focus of this work to accelerate this analysis with the use of deep learning. The proposed approach is based on deep CNNs and incorporates semantic segmentation and provides a longitudinal relationship for the same subject. The proposed approach is based on deep CNNs and incorporates semantic segmentation and provides a longitudinal relationship for the same subject. The state of art using 3D patches as inputs to modified Unet provides results around ${0.91 \pm 0.5}$ Dice and using multi-view atlas in CNNs provide around the same results. In this work, different models are explored, each offers better accuracy and fast results while increasing the segmentation quality. These methods are evaluated on 135 scans from the EADC-ADNI Harmonized Hippocampus Protocol. Proposed CNN based segmentation approaches demonstrate how 2D segmentation using prior slices can provide similar results to 3D segmentation while maintaining good continuity in the 3D dimension and improved speed. Just using 2D modified sagittal slices provide us a better Dice and longitudinal analysis for a given subject. For the ADNI dataset, using the simple UNet CNN technique gives us ${0.84 \pm 0.5}$ and while using modified CNN techniques on the same input yields ${0.89 \pm 0.5}$. Rate of atrophy and RMS error are calculated for several test cases using various methods and analyzed.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge