Jason Dowling
Deformation-aware GAN for Medical Image Synthesis with Substantially Misaligned Pairs
Aug 18, 2024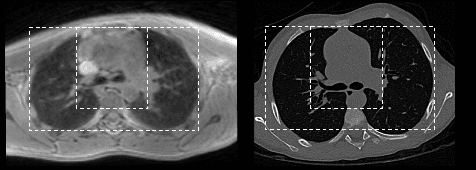
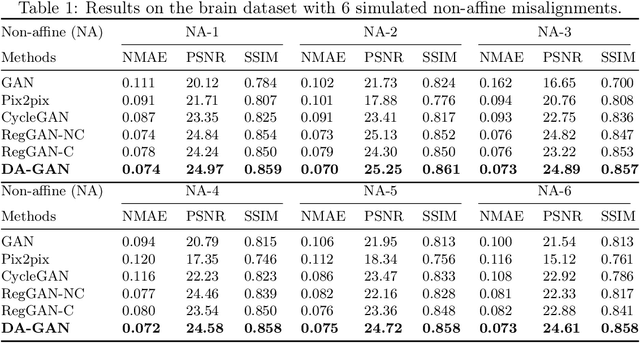
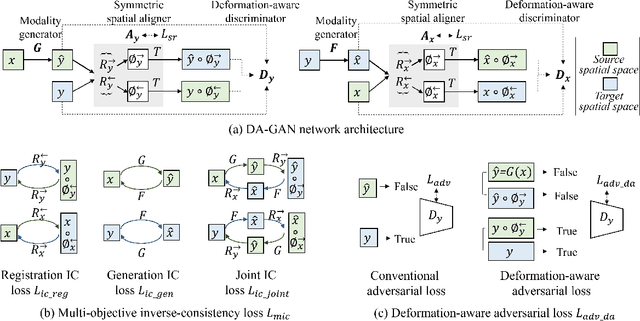
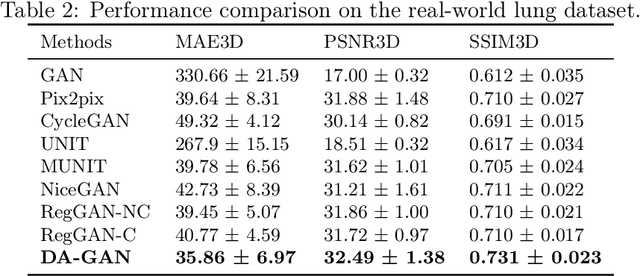
Abstract:Medical image synthesis generates additional imaging modalities that are costly, invasive or harmful to acquire, which helps to facilitate the clinical workflow. When training pairs are substantially misaligned (e.g., lung MRI-CT pairs with respiratory motion), accurate image synthesis remains a critical challenge. Recent works explored the directional registration module to adjust misalignment in generative adversarial networks (GANs); however, substantial misalignment will lead to 1) suboptimal data mapping caused by correspondence ambiguity, and 2) degraded image fidelity caused by morphology influence on discriminators. To address the challenges, we propose a novel Deformation-aware GAN (DA-GAN) to dynamically correct the misalignment during the image synthesis based on multi-objective inverse consistency. Specifically, in the generative process, three levels of inverse consistency cohesively optimise symmetric registration and image generation for improved correspondence. In the adversarial process, to further improve image fidelity under misalignment, we design deformation-aware discriminators to disentangle the mismatched spatial morphology from the judgement of image fidelity. Experimental results show that DA-GAN achieved superior performance on a public dataset with simulated misalignments and a real-world lung MRI-CT dataset with respiratory motion misalignment. The results indicate the potential for a wide range of medical image synthesis tasks such as radiotherapy planning.
e-Health CSIRO at RRG24: Entropy-Augmented Self-Critical Sequence Training for Radiology Report Generation
Aug 07, 2024Abstract:The Shared Task on Large-Scale Radiology Report Generation (RRG24) aims to expedite the development of assistive systems for interpreting and reporting on chest X-ray (CXR) images. This task challenges participants to develop models that generate the findings and impression sections of radiology reports from CXRs from a patient's study, using five different datasets. This paper outlines the e-Health CSIRO team's approach, which achieved multiple first-place finishes in RRG24. The core novelty of our approach lies in the addition of entropy regularisation to self-critical sequence training, to maintain a higher entropy in the token distribution. This prevents overfitting to common phrases and ensures a broader exploration of the vocabulary during training, essential for handling the diversity of the radiology reports in the RRG24 datasets. Our model is available on Hugging Face https://huggingface.co/aehrc/cxrmate-rrg24.
e-Health CSIRO at "Discharge Me!" 2024: Generating Discharge Summary Sections with Fine-tuned Language Models
Jul 03, 2024Abstract:Clinical documentation is an important aspect of clinicians' daily work and often demands a significant amount of time. The BioNLP 2024 Shared Task on Streamlining Discharge Documentation (Discharge Me!) aims to alleviate this documentation burden by automatically generating discharge summary sections, including brief hospital course and discharge instruction, which are often time-consuming to synthesize and write manually. We approach the generation task by fine-tuning multiple open-sourced language models (LMs), including both decoder-only and encoder-decoder LMs, with various configurations on input context. We also examine different setups for decoding algorithms, model ensembling or merging, and model specialization. Our results show that conditioning on the content of discharge summary prior to the target sections is effective for the generation task. Furthermore, we find that smaller encoder-decoder LMs can work as well or even slightly better than larger decoder based LMs fine-tuned through LoRA. The model checkpoints from our team (aehrc) are openly available.
The Impact of Auxiliary Patient Data on Automated Chest X-Ray Report Generation and How to Incorporate It
Jun 19, 2024Abstract:This study investigates the integration of diverse patient data sources into multimodal language models for automated chest X-ray (CXR) report generation. Traditionally, CXR report generation relies solely on CXR images and limited radiology data, overlooking valuable information from patient health records, particularly from emergency departments. Utilising the MIMIC-CXR and MIMIC-IV-ED datasets, we incorporate detailed patient information such as aperiodic vital signs, medications, and clinical history to enhance diagnostic accuracy. We introduce a novel approach to transform these heterogeneous data sources into embeddings that prompt a multimodal language model, significantly enhancing the diagnostic accuracy of generated radiology reports. Our comprehensive evaluation demonstrates the benefits of using a broader set of patient data, underscoring the potential for enhanced diagnostic capabilities and better patient outcomes through the integration of multimodal data in CXR report generation.
Evaluating the Impact of Sequence Combinations on Breast Tumor Segmentation in Multiparametric MRI
Jun 12, 2024Abstract:Multiparametric magnetic resonance imaging (mpMRI) is a key tool for assessing breast cancer progression. Although deep learning has been applied to automate tumor segmentation in breast MRI, the effect of sequence combinations in mpMRI remains under-investigated. This study explores the impact of different combinations of T2-weighted (T2w), dynamic contrast-enhanced MRI (DCE-MRI) and diffusion-weighted imaging (DWI) with apparent diffusion coefficient (ADC) map on breast tumor segmentation using nnU-Net. Evaluated on a multicenter mpMRI dataset, the nnU-Net model using DCE sequences achieved a Dice similarity coefficient (DSC) of 0.69 $\pm$ 0.18 for functional tumor volume (FTV) segmentation. For whole tumor mask (WTM) segmentation, adding the predicted FTV to DWI and ADC map improved the DSC from 0.57 $\pm$ 0.24 to 0.60 $\pm$ 0.21. Adding T2w did not yield significant improvement, which still requires further investigation under a more standardized imaging protocol. This study serves as a foundation for future work on predicting breast cancer treatment response using mpMRI.
Longitudinal Data and a Semantic Similarity Reward for Chest X-Ray Report Generation
Jul 19, 2023


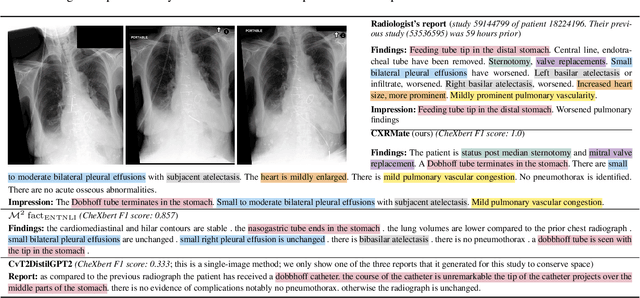
Abstract:Chest X-Ray (CXR) report generation is a promising approach to improving the efficiency of CXR interpretation. However, a significant increase in diagnostic accuracy is required before that can be realised. Motivated by this, we propose a framework that is more inline with a radiologist's workflow by considering longitudinal data. Here, the decoder is additionally conditioned on the report from the subject's previous imaging study via a prompt. We also propose a new reward for reinforcement learning based on CXR-BERT, which computes the similarity between reports. We conduct experiments on the MIMIC-CXR dataset. The results indicate that longitudinal data improves CXR report generation. CXR-BERT is also shown to be a promising alternative to the current state-of-the-art reward based on RadGraph. This investigation indicates that longitudinal CXR report generation can offer a substantial increase in diagnostic accuracy. Our Hugging Face model is available at: https://huggingface.co/aehrc/cxrmate and code is available at: https://github.com/aehrc/cxrmate.
Improving Chest X-Ray Report Generation by Leveraging Warm-Starting
Jan 24, 2022



Abstract:Automatically generating a report from a patient's Chest X-Rays (CXRs) is a promising solution to reducing clinical workload and improving patient care. However, current CXR report generators, which are predominantly encoder-to-decoder models, lack the diagnostic accuracy to be deployed in a clinical setting. To improve CXR report generation, we investigate warm-starting the encoder and decoder with recent open-source computer vision and natural language processing checkpoints, such as the Vision Transformer (ViT) and PubMedBERT. To this end, each checkpoint is evaluated on the MIMIC-CXR and IU X-Ray datasets using natural language generation and Clinical Efficacy (CE) metrics. Our experimental investigation demonstrates that the Convolutional vision Transformer (CvT) ImageNet-21K and the Distilled Generative Pre-trained Transformer 2 (DistilGPT2) checkpoints are best for warm-starting the encoder and decoder, respectively. Compared to the state-of-the-art (M2 Transformer Progressive), CvT2DistilGPT2 attained an improvement of 8.3% for CE F-1, 1.8% for BLEU-4, 1.6% for ROUGE-L, and 1.0% for METEOR. The reports generated by CvT2DistilGPT2 are more diagnostically accurate and have a higher similarity to radiologist reports than previous approaches. By leveraging warm-starting, CvT2DistilGPT2 brings automatic CXR report generation one step closer to the clinical setting. CvT2DistilGPT2 and its MIMIC-CXR checkpoint are available at https://github.com/aehrc/cvt2distilgpt2.
Going deeper with brain morphometry using neural networks
Sep 07, 2020
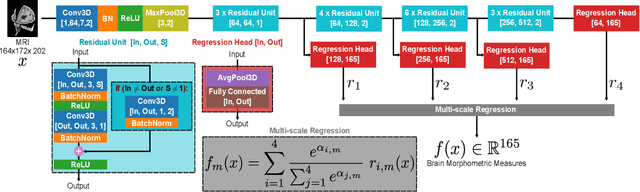
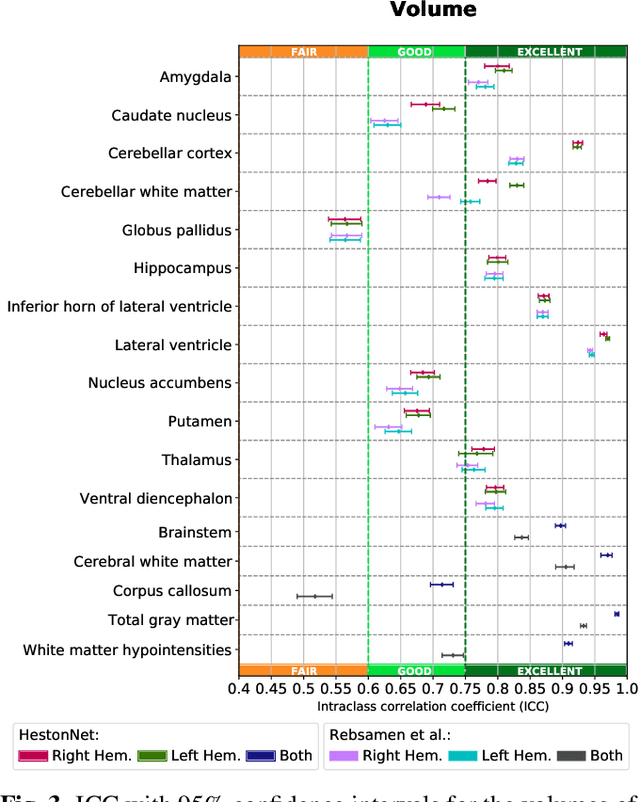
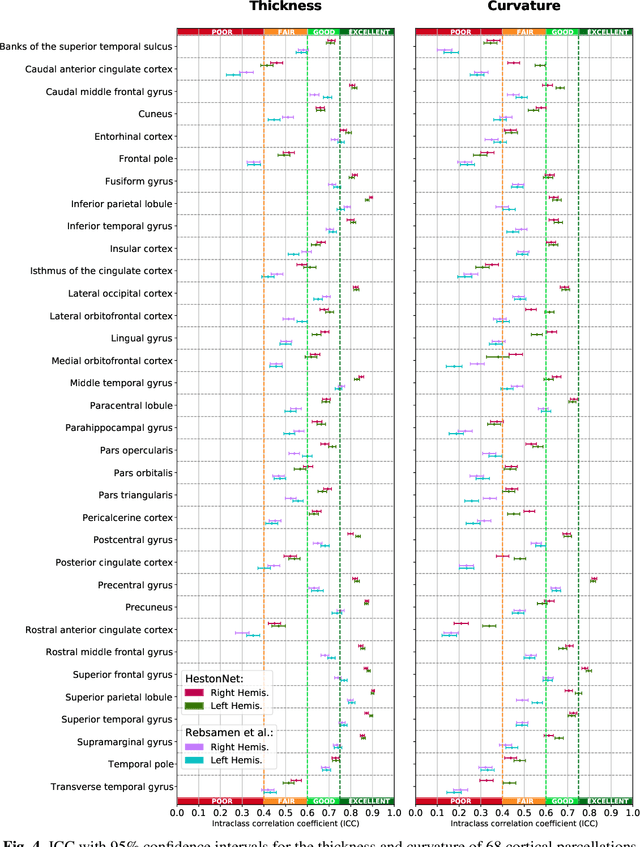
Abstract:Brain morphometry from magnetic resonance imaging (MRI) is a consolidated biomarker for many neurodegenerative diseases. Recent advances in this domain indicate that deep convolutional neural networks can infer morphometric measurements within a few seconds. Nevertheless, the accuracy of the devised model for insightful bio-markers (mean curvature and thickness) remains unsatisfactory. In this paper, we propose a more accurate and efficient neural network model for brain morphometry named HerstonNet. More specifically, we develop a 3D ResNet-based neural network to learn rich features directly from MRI, design a multi-scale regression scheme by predicting morphometric measures at feature maps of different resolutions, and leverage a robust optimization method to avoid poor quality minima and reduce the prediction variance. As a result, HerstonNet improves the existing approach by 24.30% in terms of intraclass correlation coefficient (agreement measure) to FreeSurfer silver-standards while maintaining a competitive run-time.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge