Hang Min
Standardized Evaluation of Automatic Methods for Perivascular Spaces Segmentation in MRI -- MICCAI 2024 Challenge Results
Dec 20, 2025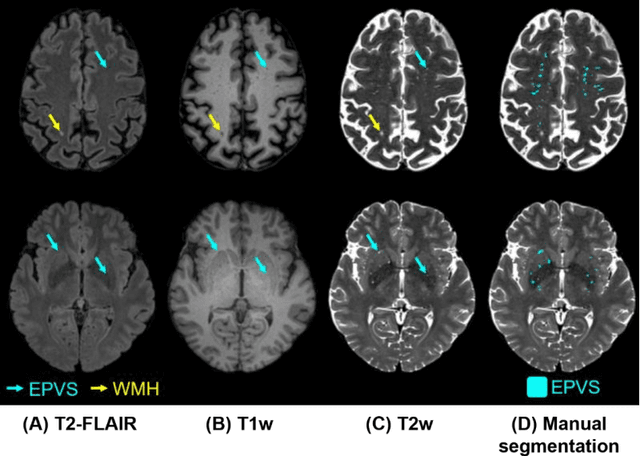
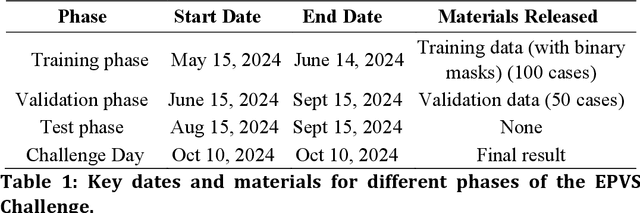
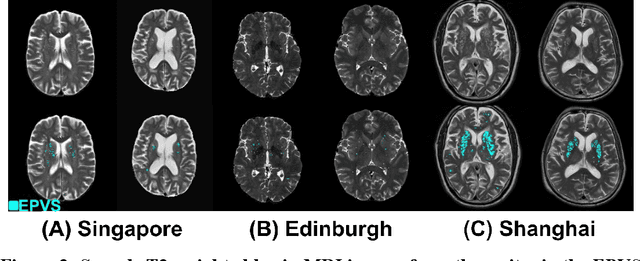
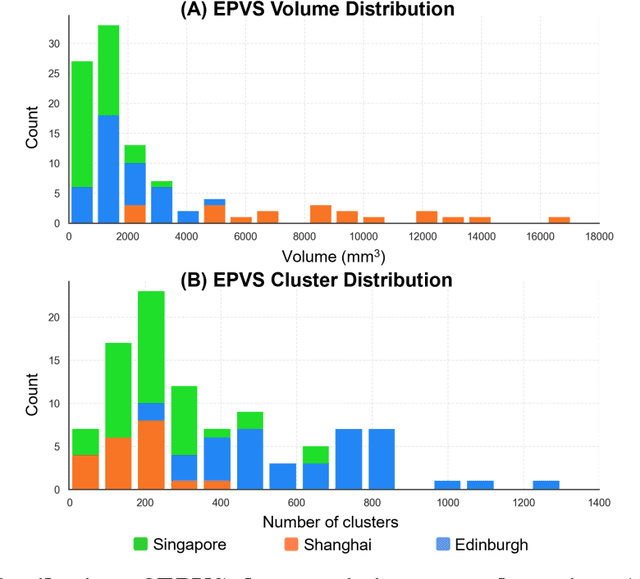
Abstract:Perivascular spaces (PVS), when abnormally enlarged and visible in magnetic resonance imaging (MRI) structural sequences, are important imaging markers of cerebral small vessel disease and potential indicators of neurodegenerative conditions. Despite their clinical significance, automatic enlarged PVS (EPVS) segmentation remains challenging due to their small size, variable morphology, similarity with other pathological features, and limited annotated datasets. This paper presents the EPVS Challenge organized at MICCAI 2024, which aims to advance the development of automated algorithms for EPVS segmentation across multi-site data. We provided a diverse dataset comprising 100 training, 50 validation, and 50 testing scans collected from multiple international sites (UK, Singapore, and China) with varying MRI protocols and demographics. All annotations followed the STRIVE protocol to ensure standardized ground truth and covered the full brain parenchyma. Seven teams completed the full challenge, implementing various deep learning approaches primarily based on U-Net architectures with innovations in multi-modal processing, ensemble strategies, and transformer-based components. Performance was evaluated using dice similarity coefficient, absolute volume difference, recall, and precision metrics. The winning method employed MedNeXt architecture with a dual 2D/3D strategy for handling varying slice thicknesses. The top solutions showed relatively good performance on test data from seen datasets, but significant degradation of performance was observed on the previously unseen Shanghai cohort, highlighting cross-site generalization challenges due to domain shift. This challenge establishes an important benchmark for EPVS segmentation methods and underscores the need for the continued development of robust algorithms that can generalize in diverse clinical settings.
Evaluating the Impact of Sequence Combinations on Breast Tumor Segmentation in Multiparametric MRI
Jun 12, 2024Abstract:Multiparametric magnetic resonance imaging (mpMRI) is a key tool for assessing breast cancer progression. Although deep learning has been applied to automate tumor segmentation in breast MRI, the effect of sequence combinations in mpMRI remains under-investigated. This study explores the impact of different combinations of T2-weighted (T2w), dynamic contrast-enhanced MRI (DCE-MRI) and diffusion-weighted imaging (DWI) with apparent diffusion coefficient (ADC) map on breast tumor segmentation using nnU-Net. Evaluated on a multicenter mpMRI dataset, the nnU-Net model using DCE sequences achieved a Dice similarity coefficient (DSC) of 0.69 $\pm$ 0.18 for functional tumor volume (FTV) segmentation. For whole tumor mask (WTM) segmentation, adding the predicted FTV to DWI and ADC map improved the DSC from 0.57 $\pm$ 0.24 to 0.60 $\pm$ 0.21. Adding T2w did not yield significant improvement, which still requires further investigation under a more standardized imaging protocol. This study serves as a foundation for future work on predicting breast cancer treatment response using mpMRI.
Automatic lesion detection, segmentation and characterization via 3D multiscale morphological sifting in breast MRI
Jul 07, 2020



Abstract:Previous studies on computer aided detection/diagnosis (CAD) in 4D breast magnetic resonance imaging (MRI) regard lesion detection, segmentation and characterization as separate tasks, and typically require users to manually select 2D MRI slices or regions of interest as the input. In this work, we present a breast MRI CAD system that can handle 4D multimodal breast MRI data, and integrate lesion detection, segmentation and characterization with no user intervention. The proposed CAD system consists of three major stages: region candidate generation, feature extraction and region candidate classification. Breast lesions are firstly extracted as region candidates using the novel 3D multiscale morphological sifting (MMS). The 3D MMS, which uses linear structuring elements to extract lesion-like patterns, can segment lesions from breast images accurately and efficiently. Analytical features are then extracted from all available 4D multimodal breast MRI sequences, including T1-, T2-weighted and DCE sequences, to represent the signal intensity, texture, morphological and enhancement kinetic characteristics of the region candidates. The region candidates are lastly classified as lesion or normal tissue by the random under-sampling boost (RUSboost), and as malignant or benign lesion by the random forest. Evaluated on a breast MRI dataset which contains a total of 117 cases with 95 malignant and 46 benign lesions, the proposed system achieves a true positive rate (TPR) of 0.90 at 3.19 false positives per patient (FPP) for lesion detection and a TPR of 0.91 at a FPP of 2.95 for identifying malignant lesions without any user intervention. The average dice similarity index (DSI) is 0.72 for lesion segmentation. Compared with previously proposed systems evaluated on the same breast MRI dataset, the proposed CAD system achieves a favourable performance in breast lesion detection and characterization.
Fully automatic computer-aided mass detection and segmentation via pseudo-color mammograms and Mask R-CNN
Jun 28, 2019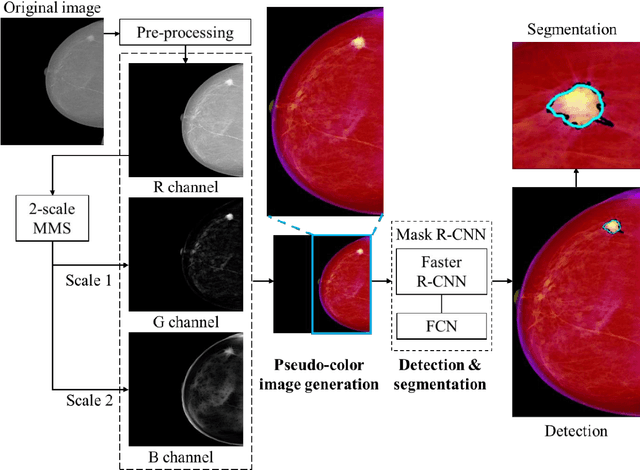

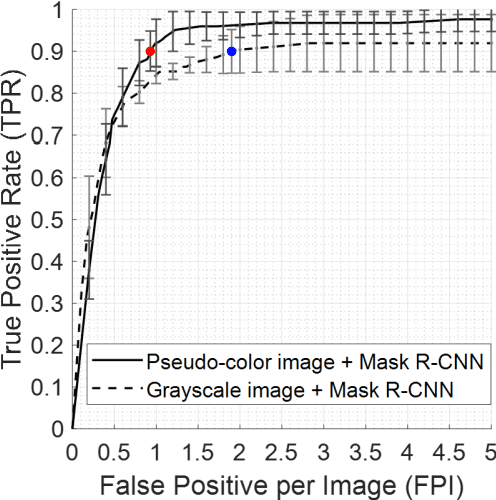

Abstract:Purpose: To propose pseudo-color mammograms that enhance mammographic masses as part of a fast computer-aided detection (CAD) system that simultaneously detects and segments masses without any user intervention. Methods: The proposed pseudo-color mammograms, whose three channels contain the original grayscale mammogram and two morphologically enhanced images, are used to provide pseudo-color contrast to the lesions. The morphological enhancement 'sifts' out the mass-like mammographic patterns to improve detection and segmentation. We construct a fast, fully automated simultaneous mass detection and segmentation CAD system using the colored mammograms as inputs of transfer learning with the Mask R-CNN which is a state-of-the-art deep learning framework. The source code for this work has been made available online. Results: Evaluated on the publicly available mammographic dataset INbreast, the method outperforms the state-of-the-art methods by achieving an average true positive rate of 0.90 at 0.9 false positive per image and an average Dice similarity index for mass segmentation of 0.88, while taking 20.4 seconds to process each image on average. Conclusions: The proposed method provides an accurate, fully-automatic breast mass detection and segmentation result in less than half a minute without any user intervention while outperforming state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge