Anthony Nguyen
Intra-Layer Recurrence in Transformers for Language Modeling
May 03, 2025Abstract:Transformer models have established new benchmarks in natural language processing; however, their increasing depth results in substantial growth in parameter counts. While existing recurrent transformer methods address this issue by reprocessing layers multiple times, they often apply recurrence indiscriminately across entire blocks of layers. In this work, we investigate Intra-Layer Recurrence (ILR), a more targeted approach that applies recurrence selectively to individual layers within a single forward pass. Our experiments show that allocating more iterations to earlier layers yields optimal results. These findings suggest that ILR offers a promising direction for optimizing recurrent structures in transformer architectures.
Rephrasing Electronic Health Records for Pretraining Clinical Language Models
Nov 28, 2024Abstract:Clinical language models are important for many applications in healthcare, but their development depends on access to extensive clinical text for pretraining. However, obtaining clinical notes from electronic health records (EHRs) at scale is challenging due to patient privacy concerns. In this study, we rephrase existing clinical notes using LLMs to generate synthetic pretraining corpora, drawing inspiration from previous work on rephrasing web data. We examine four popular small-sized LLMs (<10B) to create synthetic clinical text to pretrain both decoder-based and encoder-based language models. The method yields better results in language modeling and downstream tasks than previous synthesis approaches without referencing real clinical text. We find that augmenting original clinical notes with synthetic corpora from different LLMs improves performances even at a small token budget, showing the potential of this method to support pretraining at the institutional level or be scaled to synthesize large-scale clinical corpora.
Improved GUI Grounding via Iterative Narrowing
Nov 18, 2024



Abstract:GUI grounding, the task of identifying a precise location on an interface image from a natural language query, plays a crucial role in enhancing the capabilities of Vision-Language Model (VLM) agents. While general VLMs, such as GPT-4V, demonstrate strong performance across various tasks, their proficiency in GUI grounding remains suboptimal. Recent studies have focused on fine-tuning these models specifically for one-shot GUI grounding, yielding significant improvements over baseline performance. We introduce a visual prompting framework called Iterative Narrowing (IN) to further enhance the performance of both general and fine-tuned models in GUI grounding. For evaluation, we tested our method on a comprehensive benchmark comprising different UI platforms.
e-Health CSIRO at RRG24: Entropy-Augmented Self-Critical Sequence Training for Radiology Report Generation
Aug 07, 2024Abstract:The Shared Task on Large-Scale Radiology Report Generation (RRG24) aims to expedite the development of assistive systems for interpreting and reporting on chest X-ray (CXR) images. This task challenges participants to develop models that generate the findings and impression sections of radiology reports from CXRs from a patient's study, using five different datasets. This paper outlines the e-Health CSIRO team's approach, which achieved multiple first-place finishes in RRG24. The core novelty of our approach lies in the addition of entropy regularisation to self-critical sequence training, to maintain a higher entropy in the token distribution. This prevents overfitting to common phrases and ensures a broader exploration of the vocabulary during training, essential for handling the diversity of the radiology reports in the RRG24 datasets. Our model is available on Hugging Face https://huggingface.co/aehrc/cxrmate-rrg24.
e-Health CSIRO at "Discharge Me!" 2024: Generating Discharge Summary Sections with Fine-tuned Language Models
Jul 03, 2024Abstract:Clinical documentation is an important aspect of clinicians' daily work and often demands a significant amount of time. The BioNLP 2024 Shared Task on Streamlining Discharge Documentation (Discharge Me!) aims to alleviate this documentation burden by automatically generating discharge summary sections, including brief hospital course and discharge instruction, which are often time-consuming to synthesize and write manually. We approach the generation task by fine-tuning multiple open-sourced language models (LMs), including both decoder-only and encoder-decoder LMs, with various configurations on input context. We also examine different setups for decoding algorithms, model ensembling or merging, and model specialization. Our results show that conditioning on the content of discharge summary prior to the target sections is effective for the generation task. Furthermore, we find that smaller encoder-decoder LMs can work as well or even slightly better than larger decoder based LMs fine-tuned through LoRA. The model checkpoints from our team (aehrc) are openly available.
Improving Text-based Early Prediction by Distillation from Privileged Time-Series Text
Jan 26, 2023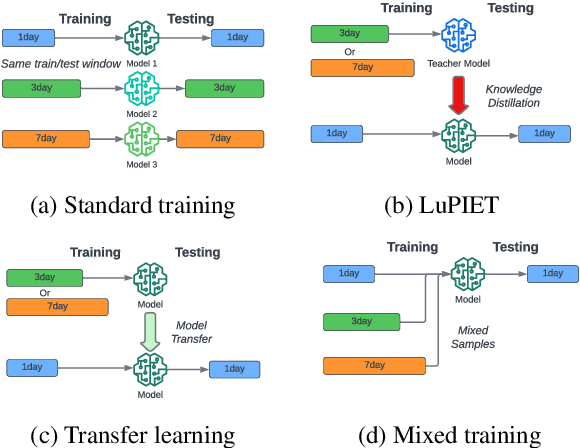
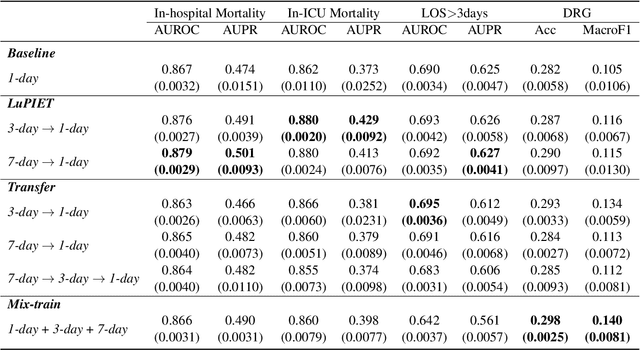
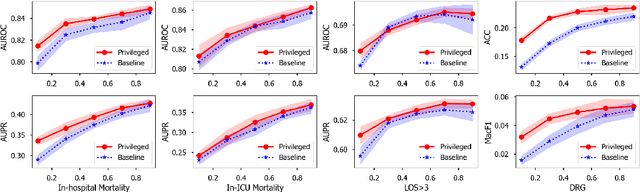
Abstract:Modeling text-based time-series to make prediction about a future event or outcome is an important task with a wide range of applications. The standard approach is to train and test the model using the same input window, but this approach neglects the data collected in longer input windows between the prediction time and the final outcome, which are often available during training. In this study, we propose to treat this neglected text as privileged information available during training to enhance early prediction modeling through knowledge distillation, presented as Learning using Privileged tIme-sEries Text (LuPIET). We evaluate the method on clinical and social media text, with four clinical prediction tasks based on clinical notes and two mental health prediction tasks based on social media posts. Our results show LuPIET is effective in enhancing text-based early predictions, though one may need to consider choosing the appropriate text representation and windows for privileged text to achieve optimal performance. Compared to two other methods using transfer learning and mixed training, LuPIET offers more stable improvements over the baseline, standard training. As far as we are concerned, this is the first study to examine learning using privileged information for time-series in the NLP context.
Automated ICD Coding using Extreme Multi-label Long Text Transformer-based Models
Dec 13, 2022



Abstract:Background: Encouraged by the success of pretrained Transformer models in many natural language processing tasks, their use for International Classification of Diseases (ICD) coding tasks is now actively being explored. In this study, we investigate three types of Transformer-based models, aiming to address the extreme label set and long text classification challenges that are posed by automated ICD coding tasks. Methods: The Transformer-based model PLM-ICD achieved the current state-of-the-art (SOTA) performance on the ICD coding benchmark dataset MIMIC-III. It was chosen as our baseline model to be further optimised. XR-Transformer, the new SOTA model in the general extreme multi-label text classification domain, and XR-LAT, a novel adaptation of the XR-Transformer model, were also trained on the MIMIC-III dataset. XR-LAT is a recursively trained model chain on a predefined hierarchical code tree with label-wise attention, knowledge transferring and dynamic negative sampling mechanisms. Results: Our optimised PLM-ICD model, which was trained with longer total and chunk sequence lengths, significantly outperformed the current SOTA PLM-ICD model, and achieved the highest micro-F1 score of 60.8%. The XR-Transformer model, although SOTA in the general domain, did not perform well across all metrics. The best XR-LAT based model obtained results that were competitive with the current SOTA PLM-ICD model, including improving the macro-AUC by 2.1%. Conclusion: Our optimised PLM-ICD model is the new SOTA model for automated ICD coding on the MIMIC-III dataset, while our novel XR-LAT model performs competitively with the previous SOTA PLM-ICD model.
Hierarchical Label-wise Attention Transformer Model for Explainable ICD Coding
Apr 22, 2022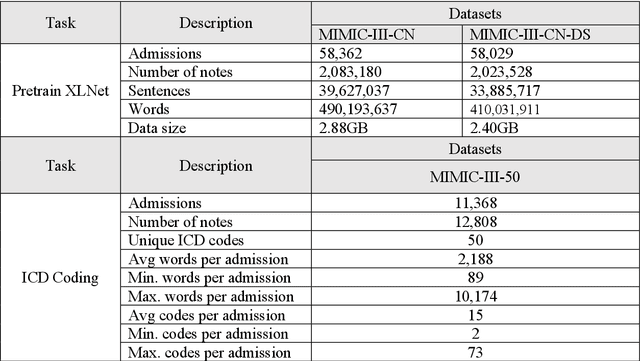
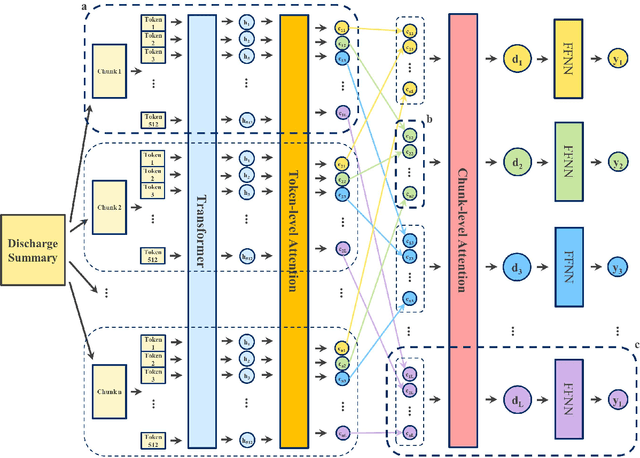
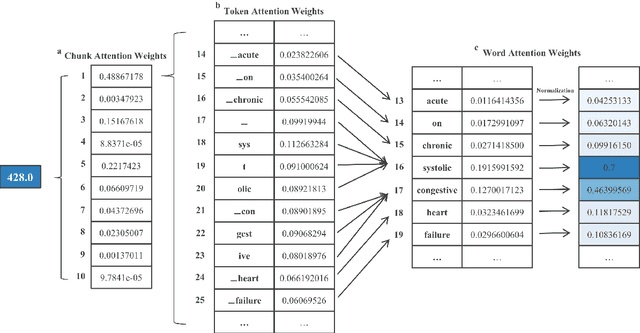
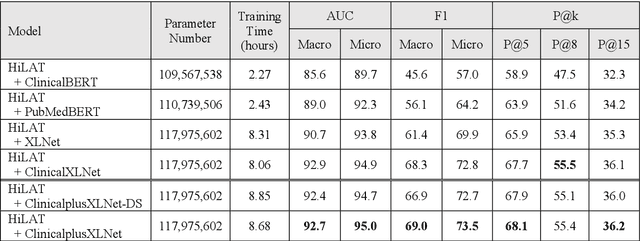
Abstract:International Classification of Diseases (ICD) coding plays an important role in systematically classifying morbidity and mortality data. In this study, we propose a hierarchical label-wise attention Transformer model (HiLAT) for the explainable prediction of ICD codes from clinical documents. HiLAT firstly fine-tunes a pretrained Transformer model to represent the tokens of clinical documents. We subsequently employ a two-level hierarchical label-wise attention mechanism that creates label-specific document representations. These representations are in turn used by a feed-forward neural network to predict whether a specific ICD code is assigned to the input clinical document of interest. We evaluate HiLAT using hospital discharge summaries and their corresponding ICD-9 codes from the MIMIC-III database. To investigate the performance of different types of Transformer models, we develop ClinicalplusXLNet, which conducts continual pretraining from XLNet-Base using all the MIMIC-III clinical notes. The experiment results show that the F1 scores of the HiLAT+ClinicalplusXLNet outperform the previous state-of-the-art models for the top-50 most frequent ICD-9 codes from MIMIC-III. Visualisations of attention weights present a potential explainability tool for checking the face validity of ICD code predictions.
De-identifying Hospital Discharge Summaries: An End-to-End Framework using Ensemble of De-Identifiers
Jan 01, 2021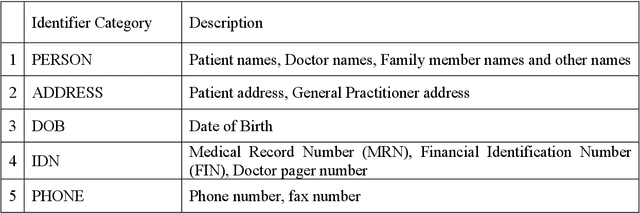
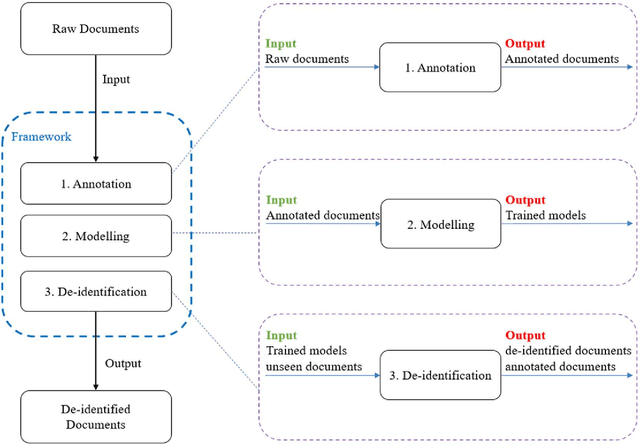
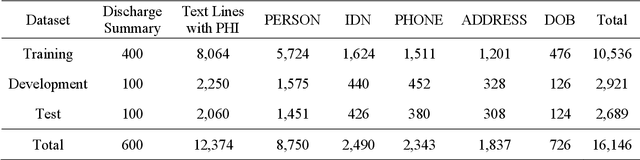
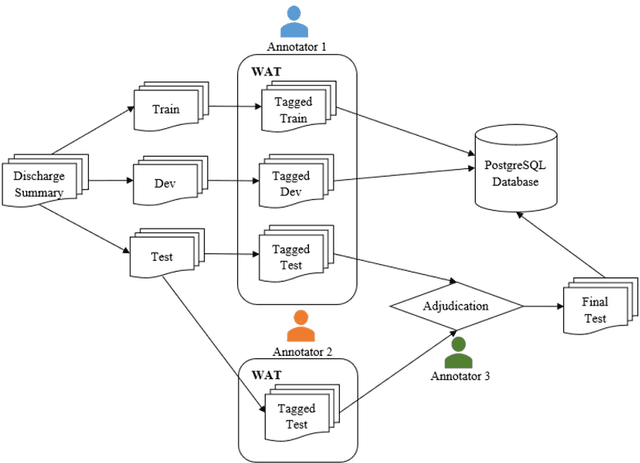
Abstract:Objective:Electronic Medical Records (EMRs) contain clinical narrative text that is of great potential value to medical researchers. However, this information is mixed with Protected Health Information (PHI) that presents risks to patient and clinician confidentiality. This paper presents an end-to-end de-identification framework to automatically remove PHI from hospital discharge summaries. Materials and Methods:Our corpus included 600 hospital discharge summaries which were extracted from the EMRs of two principal referral hospitals in Sydney, Australia. Our end-to-end de-identification framework consists of three components: 1) Annotation: labelling of PHI in the 600 hospital discharge summaries using five pre-defined categories: person, address, date of birth, individual identification number, phone/fax number; 2) Modelling: training and evaluating ensembles of named entity recognition (NER) models through the use of three natural language processing (NLP) toolkits (Stanza, FLAIR and spaCy) and both balanced and imbalanced datasets; and 3) De-identification: removing PHI from the hospital discharge summaries. Results:The final model in our framework was an ensemble which combined six single models using both balanced and imbalanced datasets for training majority voting. It achieved 0.9866 precision, 0.9862 recall and 0.9864 F1 scores. The majority of false positives and false negatives were related to the person category. Discussion:Our study showed that the ensemble of different models which were trained using three different NLP toolkits upon balanced and imbalanced datasets can achieve good results even with a relatively small corpus. Conclusion:Our end-to-end framework provides a robust solution to de-identifying clinical narrative corpuses safely. It can be easily applied to any kind of clinical narrative documents.
Stochastic Multi-level Composition Optimization Algorithms with Level-Independent Convergence Rates
Sep 07, 2020
Abstract:In this paper, we study smooth stochastic multi-level composition optimization problems, where the objective function is a nested composition of $T$ functions. We assume access to noisy evaluations of the functions and their gradients, through a stochastic first-order oracle. For solving this class of problems, we propose two algorithms using moving-average stochastic estimates, and analyze their convergence to an $\epsilon$-stationary point of the problem. We show that the first algorithm, which is a generalization of [22] to the $T$ level case, can achieve a sample complexity of $\mathcal{O}(1/\epsilon^6)$ by using mini-batches of samples in each iteration. By modifying this algorithm using linearized stochastic estimates of the function values, we improve the sample complexity to $\mathcal{O}(1/\epsilon^4)$. This modification also removes the requirement of having a mini-batch of samples in each iteration. To the best of our knowledge, this is the first time that such an online algorithm designed for the (un)constrained multi-level setting, obtains the same sample complexity of the smooth single-level setting, under mild assumptions on the stochastic first-order oracle.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge