Vicki Bennett
Automated ICD Coding using Extreme Multi-label Long Text Transformer-based Models
Dec 13, 2022



Abstract:Background: Encouraged by the success of pretrained Transformer models in many natural language processing tasks, their use for International Classification of Diseases (ICD) coding tasks is now actively being explored. In this study, we investigate three types of Transformer-based models, aiming to address the extreme label set and long text classification challenges that are posed by automated ICD coding tasks. Methods: The Transformer-based model PLM-ICD achieved the current state-of-the-art (SOTA) performance on the ICD coding benchmark dataset MIMIC-III. It was chosen as our baseline model to be further optimised. XR-Transformer, the new SOTA model in the general extreme multi-label text classification domain, and XR-LAT, a novel adaptation of the XR-Transformer model, were also trained on the MIMIC-III dataset. XR-LAT is a recursively trained model chain on a predefined hierarchical code tree with label-wise attention, knowledge transferring and dynamic negative sampling mechanisms. Results: Our optimised PLM-ICD model, which was trained with longer total and chunk sequence lengths, significantly outperformed the current SOTA PLM-ICD model, and achieved the highest micro-F1 score of 60.8%. The XR-Transformer model, although SOTA in the general domain, did not perform well across all metrics. The best XR-LAT based model obtained results that were competitive with the current SOTA PLM-ICD model, including improving the macro-AUC by 2.1%. Conclusion: Our optimised PLM-ICD model is the new SOTA model for automated ICD coding on the MIMIC-III dataset, while our novel XR-LAT model performs competitively with the previous SOTA PLM-ICD model.
Hierarchical Label-wise Attention Transformer Model for Explainable ICD Coding
Apr 22, 2022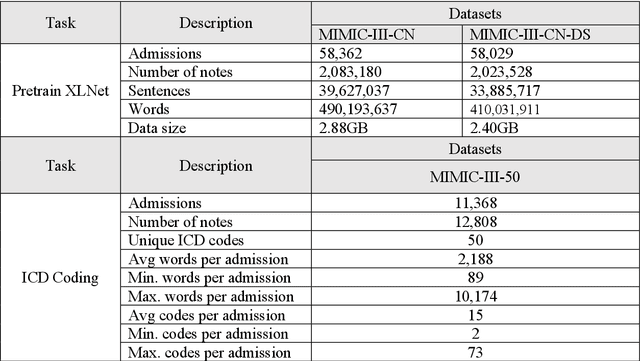
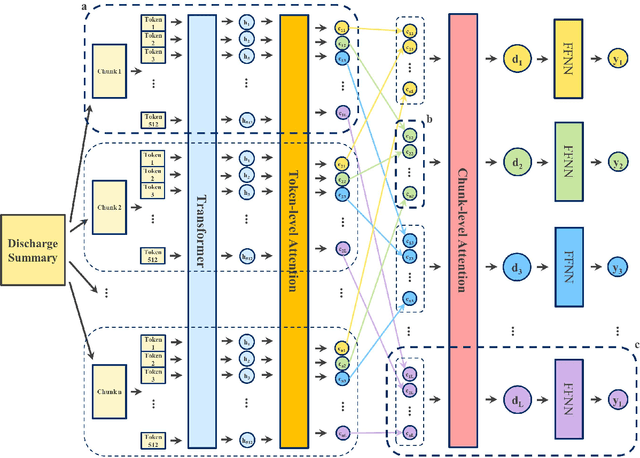
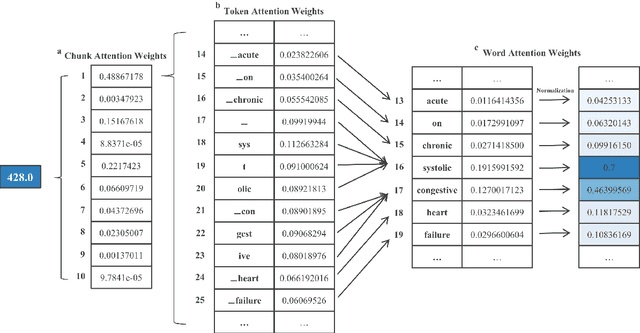
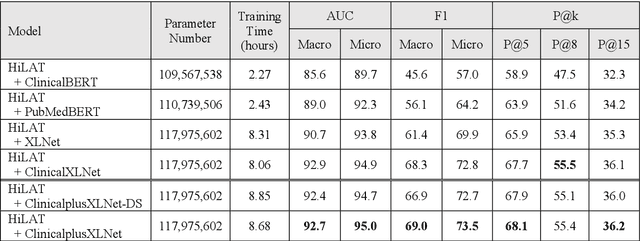
Abstract:International Classification of Diseases (ICD) coding plays an important role in systematically classifying morbidity and mortality data. In this study, we propose a hierarchical label-wise attention Transformer model (HiLAT) for the explainable prediction of ICD codes from clinical documents. HiLAT firstly fine-tunes a pretrained Transformer model to represent the tokens of clinical documents. We subsequently employ a two-level hierarchical label-wise attention mechanism that creates label-specific document representations. These representations are in turn used by a feed-forward neural network to predict whether a specific ICD code is assigned to the input clinical document of interest. We evaluate HiLAT using hospital discharge summaries and their corresponding ICD-9 codes from the MIMIC-III database. To investigate the performance of different types of Transformer models, we develop ClinicalplusXLNet, which conducts continual pretraining from XLNet-Base using all the MIMIC-III clinical notes. The experiment results show that the F1 scores of the HiLAT+ClinicalplusXLNet outperform the previous state-of-the-art models for the top-50 most frequent ICD-9 codes from MIMIC-III. Visualisations of attention weights present a potential explainability tool for checking the face validity of ICD code predictions.
De-identifying Hospital Discharge Summaries: An End-to-End Framework using Ensemble of De-Identifiers
Jan 01, 2021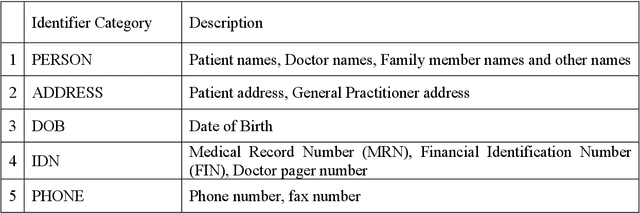
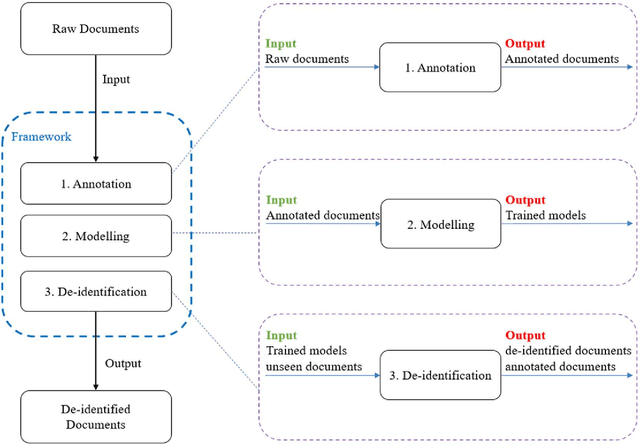
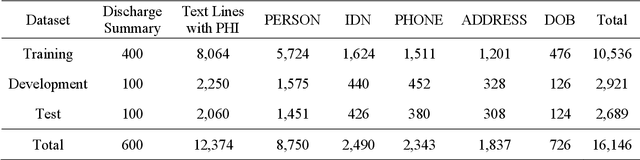
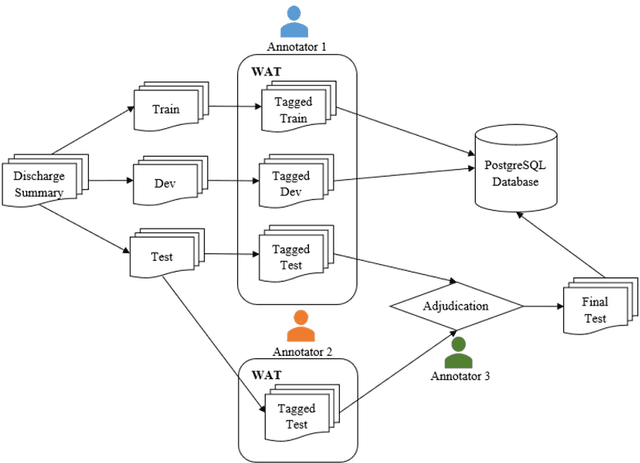
Abstract:Objective:Electronic Medical Records (EMRs) contain clinical narrative text that is of great potential value to medical researchers. However, this information is mixed with Protected Health Information (PHI) that presents risks to patient and clinician confidentiality. This paper presents an end-to-end de-identification framework to automatically remove PHI from hospital discharge summaries. Materials and Methods:Our corpus included 600 hospital discharge summaries which were extracted from the EMRs of two principal referral hospitals in Sydney, Australia. Our end-to-end de-identification framework consists of three components: 1) Annotation: labelling of PHI in the 600 hospital discharge summaries using five pre-defined categories: person, address, date of birth, individual identification number, phone/fax number; 2) Modelling: training and evaluating ensembles of named entity recognition (NER) models through the use of three natural language processing (NLP) toolkits (Stanza, FLAIR and spaCy) and both balanced and imbalanced datasets; and 3) De-identification: removing PHI from the hospital discharge summaries. Results:The final model in our framework was an ensemble which combined six single models using both balanced and imbalanced datasets for training majority voting. It achieved 0.9866 precision, 0.9862 recall and 0.9864 F1 scores. The majority of false positives and false negatives were related to the person category. Discussion:Our study showed that the ensemble of different models which were trained using three different NLP toolkits upon balanced and imbalanced datasets can achieve good results even with a relatively small corpus. Conclusion:Our end-to-end framework provides a robust solution to de-identifying clinical narrative corpuses safely. It can be easily applied to any kind of clinical narrative documents.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge