Leibo Liu
A Unified Geometric Field Theory Framework for Transformers: From Manifold Embeddings to Kernel Modulation
Nov 12, 2025Abstract:The Transformer architecture has achieved tremendous success in natural language processing, computer vision, and scientific computing through its self-attention mechanism. However, its core components-positional encoding and attention mechanisms-have lacked a unified physical or mathematical interpretation. This paper proposes a structural theoretical framework that integrates positional encoding, kernel integral operators, and attention mechanisms for in-depth theoretical investigation. We map discrete positions (such as text token indices and image pixel coordinates) to spatial functions on continuous manifolds, enabling a field-theoretic interpretation of Transformer layers as kernel-modulated operators acting over embedded manifolds.
Harmonising the Clinical Melody: Tuning Large Language Models for Hospital Course Summarisation in Clinical Coding
Sep 24, 2024Abstract:The increasing volume and complexity of clinical documentation in Electronic Medical Records systems pose significant challenges for clinical coders, who must mentally process and summarise vast amounts of clinical text to extract essential information needed for coding tasks. While large language models have been successfully applied to shorter summarisation tasks in recent years, the challenge of summarising a hospital course remains an open area for further research and development. In this study, we adapted three pre trained LLMs, Llama 3, BioMistral, Mistral Instruct v0.1 for the hospital course summarisation task, using Quantized Low Rank Adaptation fine tuning. We created a free text clinical dataset from MIMIC III data by concatenating various clinical notes as the input clinical text, paired with ground truth Brief Hospital Course sections extracted from the discharge summaries for model training. The fine tuned models were evaluated using BERTScore and ROUGE metrics to assess the effectiveness of clinical domain fine tuning. Additionally, we validated their practical utility using a novel hospital course summary assessment metric specifically tailored for clinical coding. Our findings indicate that fine tuning pre trained LLMs for the clinical domain can significantly enhance their performance in hospital course summarisation and suggest their potential as assistive tools for clinical coding. Future work should focus on refining data curation methods to create higher quality clinical datasets tailored for hospital course summary tasks and adapting more advanced open source LLMs comparable to proprietary models to further advance this research.
Automated ICD Coding using Extreme Multi-label Long Text Transformer-based Models
Dec 13, 2022



Abstract:Background: Encouraged by the success of pretrained Transformer models in many natural language processing tasks, their use for International Classification of Diseases (ICD) coding tasks is now actively being explored. In this study, we investigate three types of Transformer-based models, aiming to address the extreme label set and long text classification challenges that are posed by automated ICD coding tasks. Methods: The Transformer-based model PLM-ICD achieved the current state-of-the-art (SOTA) performance on the ICD coding benchmark dataset MIMIC-III. It was chosen as our baseline model to be further optimised. XR-Transformer, the new SOTA model in the general extreme multi-label text classification domain, and XR-LAT, a novel adaptation of the XR-Transformer model, were also trained on the MIMIC-III dataset. XR-LAT is a recursively trained model chain on a predefined hierarchical code tree with label-wise attention, knowledge transferring and dynamic negative sampling mechanisms. Results: Our optimised PLM-ICD model, which was trained with longer total and chunk sequence lengths, significantly outperformed the current SOTA PLM-ICD model, and achieved the highest micro-F1 score of 60.8%. The XR-Transformer model, although SOTA in the general domain, did not perform well across all metrics. The best XR-LAT based model obtained results that were competitive with the current SOTA PLM-ICD model, including improving the macro-AUC by 2.1%. Conclusion: Our optimised PLM-ICD model is the new SOTA model for automated ICD coding on the MIMIC-III dataset, while our novel XR-LAT model performs competitively with the previous SOTA PLM-ICD model.
HQNAS: Auto CNN deployment framework for joint quantization and architecture search
Oct 16, 2022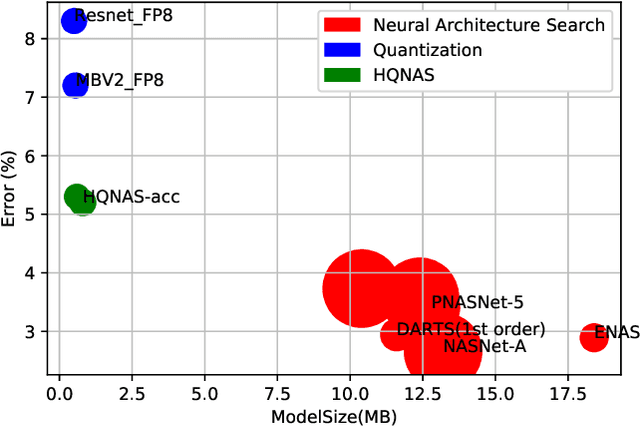
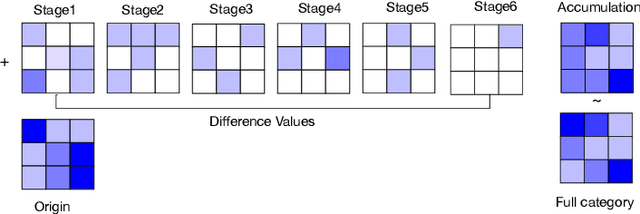
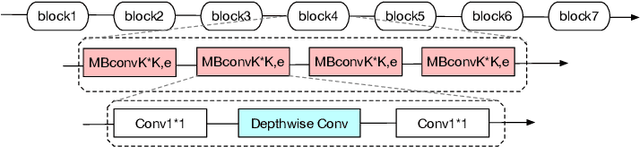
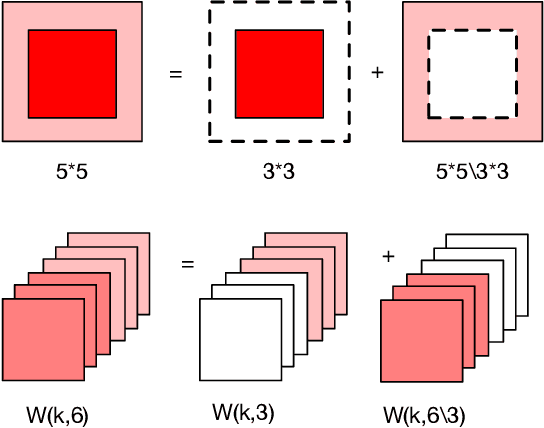
Abstract:Deep learning applications are being transferred from the cloud to edge with the rapid development of embedded computing systems. In order to achieve higher energy efficiency with the limited resource budget, neural networks(NNs) must be carefully designed in two steps, the architecture design and the quantization policy choice. Neural Architecture Search(NAS) and Quantization have been proposed separately when deploying NNs onto embedded devices. However, taking the two steps individually is time-consuming and leads to a sub-optimal final deployment. To this end, we propose a novel neural network design framework called Hardware-aware Quantized Neural Architecture Search(HQNAS) framework which combines the NAS and Quantization together in a very efficient manner using weight-sharing and bit-sharing. It takes only 4 GPU hours to discover an outstanding NN policy on CIFAR10. It also takes only %10 GPU time to generate a comparable model on Imagenet compared to the traditional NAS method with 1.8x decrease of latency and a negligible accuracy loss of only 0.7%. Besides, our method can be adapted in a lifelong situation where the neural network needs to evolve occasionally due to changes of local data, environment and user preference.
FAQS: Communication-efficient Federate DNN Architecture and Quantization Co-Search for personalized Hardware-aware Preferences
Oct 16, 2022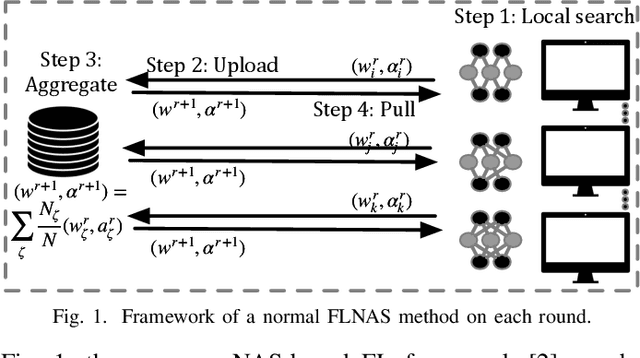
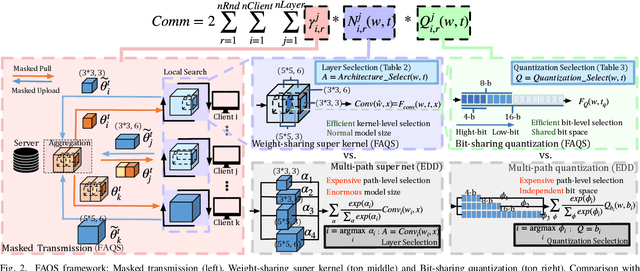
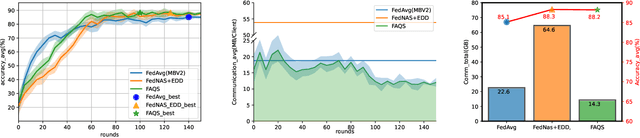
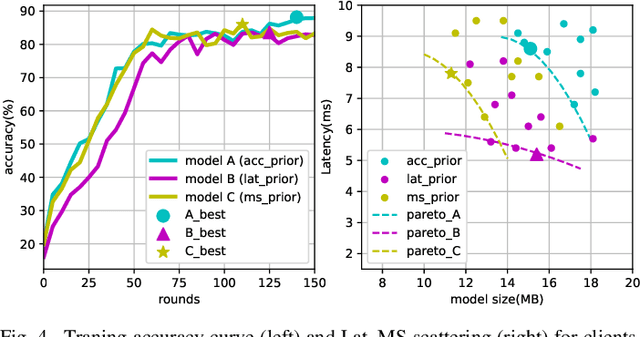
Abstract:Due to user privacy and regulatory restrictions, federate learning (FL) is proposed as a distributed learning framework for training deep neural networks (DNN) on decentralized data clients. Recent advancements in FL have applied Neural Architecture Search (NAS) to replace the predefined one-size-fit-all DNN model, which is not optimal for all tasks of various data distributions, with searchable DNN architectures. However, previous methods suffer from expensive communication cost rasied by frequent large model parameters transmission between the server and clients. Such difficulty is further amplified when combining NAS algorithms, which commonly require prohibitive computation and enormous model storage. Towards this end, we propose FAQS, an efficient personalized FL-NAS-Quantization framework to reduce the communication cost with three features: weight-sharing super kernels, bit-sharing quantization and masked transmission. FAQS has an affordable search time and demands very limited size of transmitted messages at each round. By setting different personlized pareto function loss on local clients, FAQS can yield heterogeneous hardware-aware models for various user preferences. Experimental results show that FAQS achieves average reduction of 1.58x in communication bandwith per round compared with normal FL framework and 4.51x compared with FL+NAS framwork.
Hierarchical Label-wise Attention Transformer Model for Explainable ICD Coding
Apr 22, 2022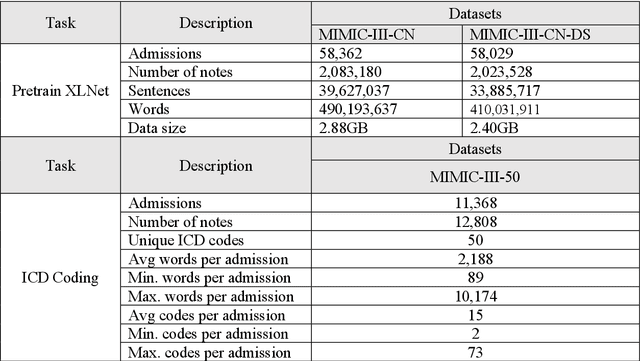
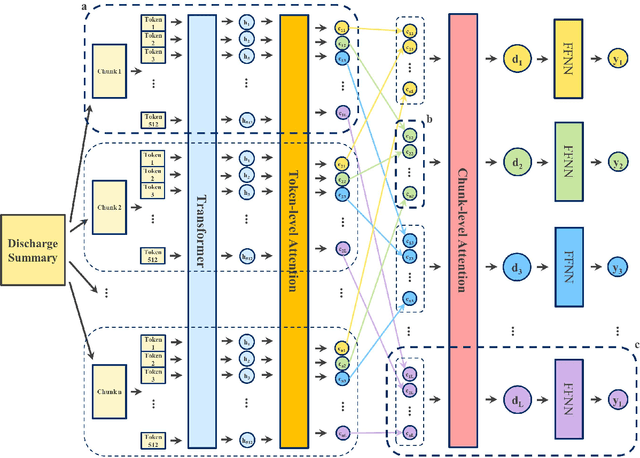
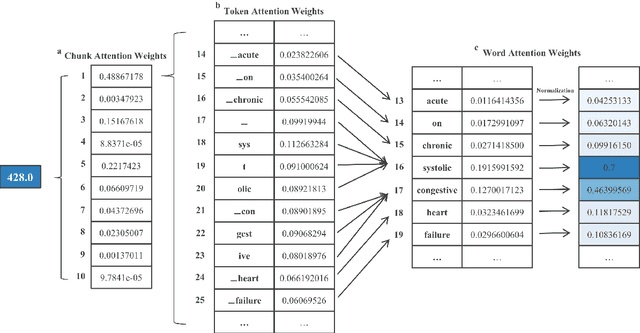
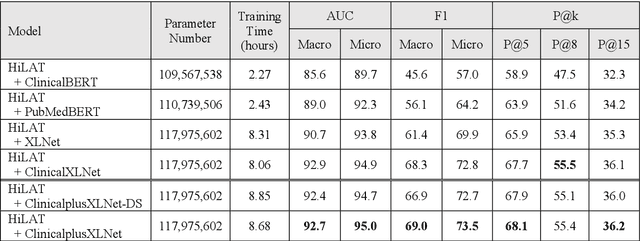
Abstract:International Classification of Diseases (ICD) coding plays an important role in systematically classifying morbidity and mortality data. In this study, we propose a hierarchical label-wise attention Transformer model (HiLAT) for the explainable prediction of ICD codes from clinical documents. HiLAT firstly fine-tunes a pretrained Transformer model to represent the tokens of clinical documents. We subsequently employ a two-level hierarchical label-wise attention mechanism that creates label-specific document representations. These representations are in turn used by a feed-forward neural network to predict whether a specific ICD code is assigned to the input clinical document of interest. We evaluate HiLAT using hospital discharge summaries and their corresponding ICD-9 codes from the MIMIC-III database. To investigate the performance of different types of Transformer models, we develop ClinicalplusXLNet, which conducts continual pretraining from XLNet-Base using all the MIMIC-III clinical notes. The experiment results show that the F1 scores of the HiLAT+ClinicalplusXLNet outperform the previous state-of-the-art models for the top-50 most frequent ICD-9 codes from MIMIC-III. Visualisations of attention weights present a potential explainability tool for checking the face validity of ICD code predictions.
De-identifying Hospital Discharge Summaries: An End-to-End Framework using Ensemble of De-Identifiers
Jan 01, 2021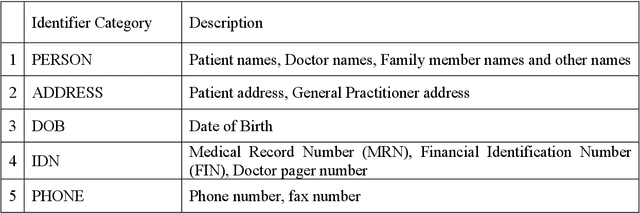
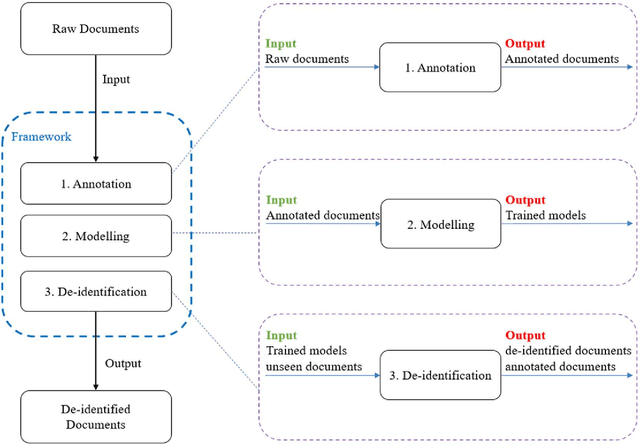
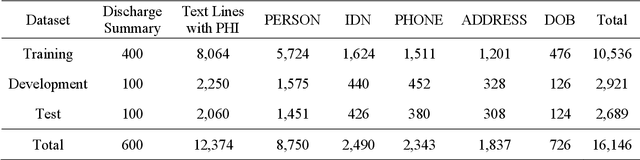
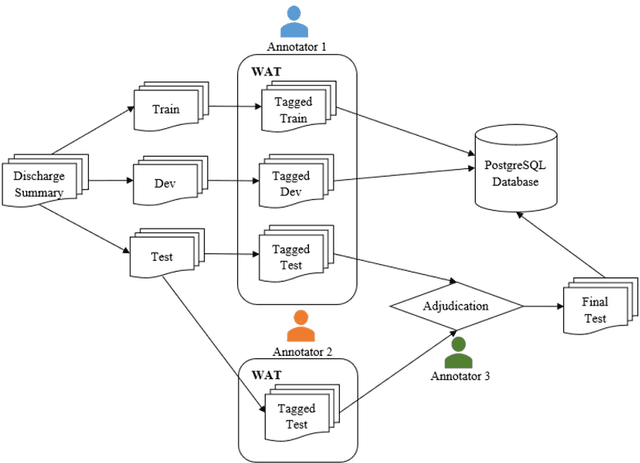
Abstract:Objective:Electronic Medical Records (EMRs) contain clinical narrative text that is of great potential value to medical researchers. However, this information is mixed with Protected Health Information (PHI) that presents risks to patient and clinician confidentiality. This paper presents an end-to-end de-identification framework to automatically remove PHI from hospital discharge summaries. Materials and Methods:Our corpus included 600 hospital discharge summaries which were extracted from the EMRs of two principal referral hospitals in Sydney, Australia. Our end-to-end de-identification framework consists of three components: 1) Annotation: labelling of PHI in the 600 hospital discharge summaries using five pre-defined categories: person, address, date of birth, individual identification number, phone/fax number; 2) Modelling: training and evaluating ensembles of named entity recognition (NER) models through the use of three natural language processing (NLP) toolkits (Stanza, FLAIR and spaCy) and both balanced and imbalanced datasets; and 3) De-identification: removing PHI from the hospital discharge summaries. Results:The final model in our framework was an ensemble which combined six single models using both balanced and imbalanced datasets for training majority voting. It achieved 0.9866 precision, 0.9862 recall and 0.9864 F1 scores. The majority of false positives and false negatives were related to the person category. Discussion:Our study showed that the ensemble of different models which were trained using three different NLP toolkits upon balanced and imbalanced datasets can achieve good results even with a relatively small corpus. Conclusion:Our end-to-end framework provides a robust solution to de-identifying clinical narrative corpuses safely. It can be easily applied to any kind of clinical narrative documents.
Small-footprint Keyword Spotting with Graph Convolutional Network
Dec 11, 2019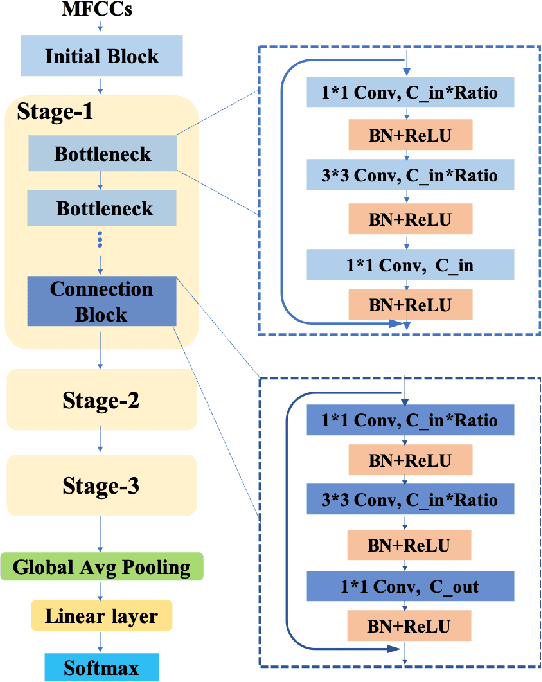
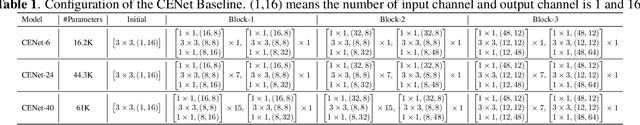
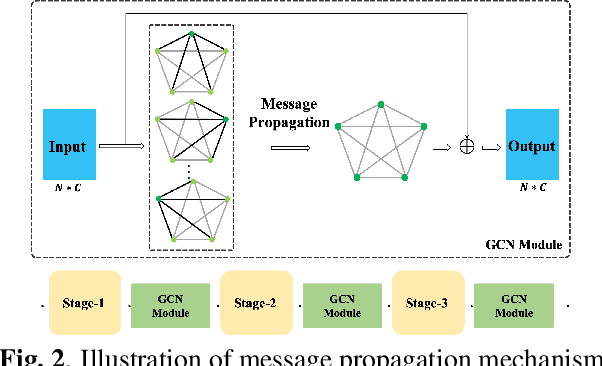
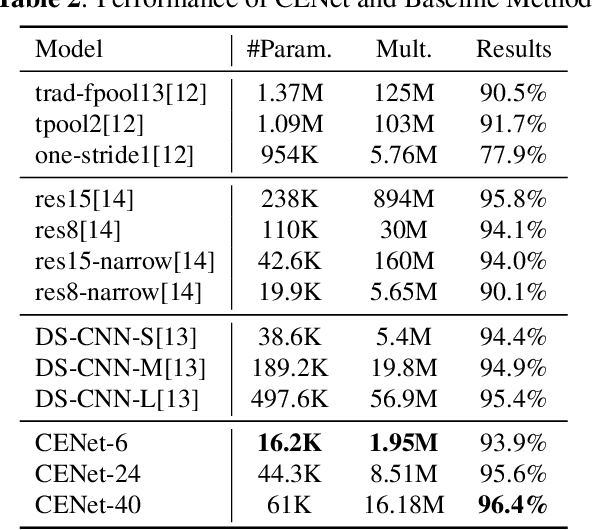
Abstract:Despite the recent successes of deep neural networks, it remains challenging to achieve high precision keyword spotting task (KWS) on resource-constrained devices. In this study, we propose a novel context-aware and compact architecture for keyword spotting task. Based on residual connection and bottleneck structure, we design a compact and efficient network for KWS task. To leverage the long range dependencies and global context of the convolutional feature maps, the graph convolutional network is introduced to encode the non-local relations. By evaluated on the Google Speech Command Dataset, the proposed method achieves state-of-the-art performance and outperforms the prior works by a large margin with lower computational cost.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge