Oscar Perez-Concha
Harmonising the Clinical Melody: Tuning Large Language Models for Hospital Course Summarisation in Clinical Coding
Sep 24, 2024Abstract:The increasing volume and complexity of clinical documentation in Electronic Medical Records systems pose significant challenges for clinical coders, who must mentally process and summarise vast amounts of clinical text to extract essential information needed for coding tasks. While large language models have been successfully applied to shorter summarisation tasks in recent years, the challenge of summarising a hospital course remains an open area for further research and development. In this study, we adapted three pre trained LLMs, Llama 3, BioMistral, Mistral Instruct v0.1 for the hospital course summarisation task, using Quantized Low Rank Adaptation fine tuning. We created a free text clinical dataset from MIMIC III data by concatenating various clinical notes as the input clinical text, paired with ground truth Brief Hospital Course sections extracted from the discharge summaries for model training. The fine tuned models were evaluated using BERTScore and ROUGE metrics to assess the effectiveness of clinical domain fine tuning. Additionally, we validated their practical utility using a novel hospital course summary assessment metric specifically tailored for clinical coding. Our findings indicate that fine tuning pre trained LLMs for the clinical domain can significantly enhance their performance in hospital course summarisation and suggest their potential as assistive tools for clinical coding. Future work should focus on refining data curation methods to create higher quality clinical datasets tailored for hospital course summary tasks and adapting more advanced open source LLMs comparable to proprietary models to further advance this research.
Continuous time recurrent neural networks: overview and application to forecasting blood glucose in the intensive care unit
Apr 14, 2023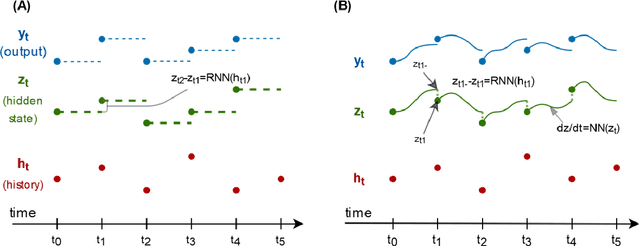
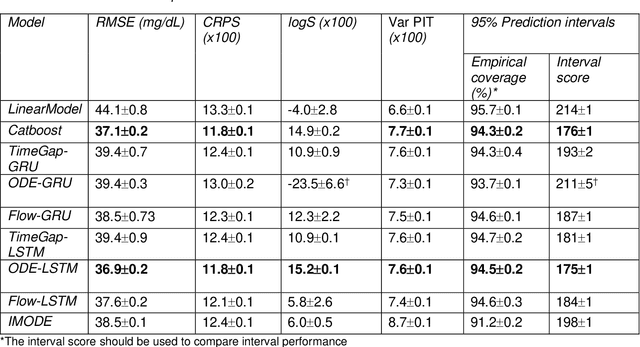
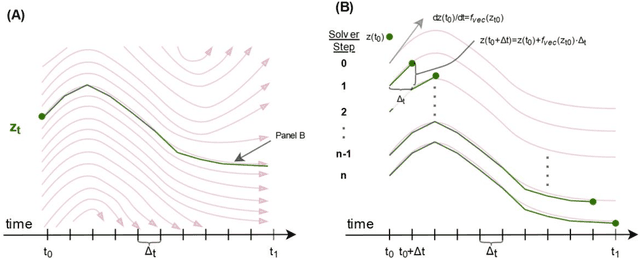
Abstract:Irregularly measured time series are common in many of the applied settings in which time series modelling is a key statistical tool, including medicine. This provides challenges in model choice, often necessitating imputation or similar strategies. Continuous time autoregressive recurrent neural networks (CTRNNs) are a deep learning model that account for irregular observations through incorporating continuous evolution of the hidden states between observations. This is achieved using a neural ordinary differential equation (ODE) or neural flow layer. In this manuscript, we give an overview of these models, including the varying architectures that have been proposed to account for issues such as ongoing medical interventions. Further, we demonstrate the application of these models to probabilistic forecasting of blood glucose in a critical care setting using electronic medical record and simulated data. The experiments confirm that addition of a neural ODE or neural flow layer generally improves the performance of autoregressive recurrent neural networks in the irregular measurement setting. However, several CTRNN architecture are outperformed by an autoregressive gradient boosted tree model (Catboost), with only a long short-term memory (LSTM) and neural ODE based architecture (ODE-LSTM) achieving comparable performance on probabilistic forecasting metrics such as the continuous ranked probability score (ODE-LSTM: 0.118$\pm$0.001; Catboost: 0.118$\pm$0.001), ignorance score (0.152$\pm$0.008; 0.149$\pm$0.002) and interval score (175$\pm$1; 176$\pm$1).
Automated ICD Coding using Extreme Multi-label Long Text Transformer-based Models
Dec 13, 2022



Abstract:Background: Encouraged by the success of pretrained Transformer models in many natural language processing tasks, their use for International Classification of Diseases (ICD) coding tasks is now actively being explored. In this study, we investigate three types of Transformer-based models, aiming to address the extreme label set and long text classification challenges that are posed by automated ICD coding tasks. Methods: The Transformer-based model PLM-ICD achieved the current state-of-the-art (SOTA) performance on the ICD coding benchmark dataset MIMIC-III. It was chosen as our baseline model to be further optimised. XR-Transformer, the new SOTA model in the general extreme multi-label text classification domain, and XR-LAT, a novel adaptation of the XR-Transformer model, were also trained on the MIMIC-III dataset. XR-LAT is a recursively trained model chain on a predefined hierarchical code tree with label-wise attention, knowledge transferring and dynamic negative sampling mechanisms. Results: Our optimised PLM-ICD model, which was trained with longer total and chunk sequence lengths, significantly outperformed the current SOTA PLM-ICD model, and achieved the highest micro-F1 score of 60.8%. The XR-Transformer model, although SOTA in the general domain, did not perform well across all metrics. The best XR-LAT based model obtained results that were competitive with the current SOTA PLM-ICD model, including improving the macro-AUC by 2.1%. Conclusion: Our optimised PLM-ICD model is the new SOTA model for automated ICD coding on the MIMIC-III dataset, while our novel XR-LAT model performs competitively with the previous SOTA PLM-ICD model.
Hierarchical Label-wise Attention Transformer Model for Explainable ICD Coding
Apr 22, 2022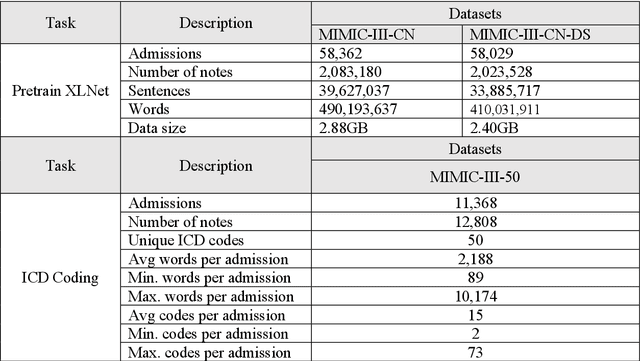
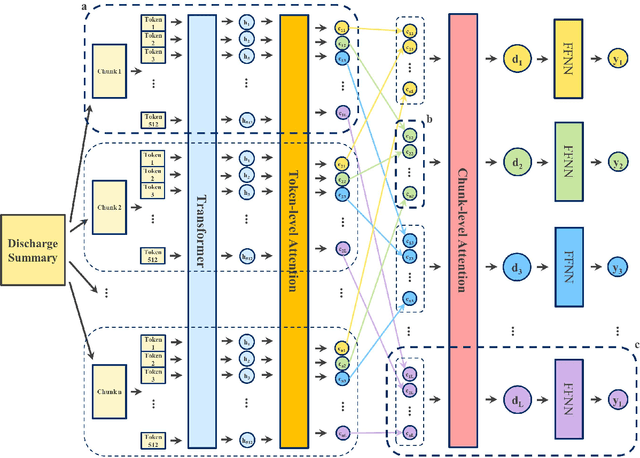
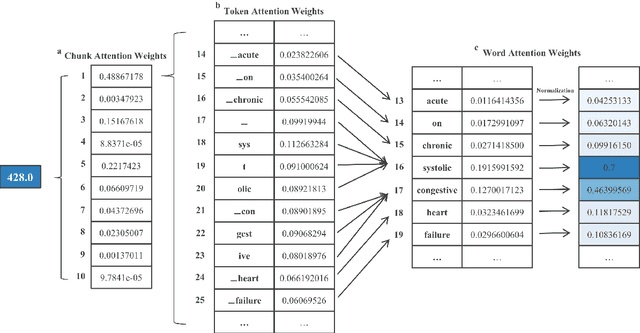
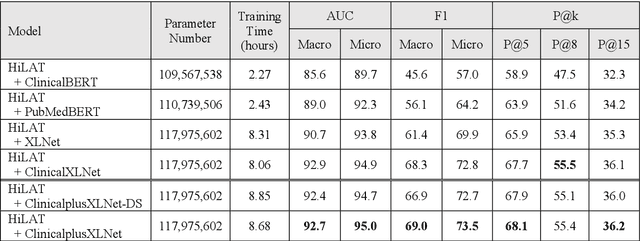
Abstract:International Classification of Diseases (ICD) coding plays an important role in systematically classifying morbidity and mortality data. In this study, we propose a hierarchical label-wise attention Transformer model (HiLAT) for the explainable prediction of ICD codes from clinical documents. HiLAT firstly fine-tunes a pretrained Transformer model to represent the tokens of clinical documents. We subsequently employ a two-level hierarchical label-wise attention mechanism that creates label-specific document representations. These representations are in turn used by a feed-forward neural network to predict whether a specific ICD code is assigned to the input clinical document of interest. We evaluate HiLAT using hospital discharge summaries and their corresponding ICD-9 codes from the MIMIC-III database. To investigate the performance of different types of Transformer models, we develop ClinicalplusXLNet, which conducts continual pretraining from XLNet-Base using all the MIMIC-III clinical notes. The experiment results show that the F1 scores of the HiLAT+ClinicalplusXLNet outperform the previous state-of-the-art models for the top-50 most frequent ICD-9 codes from MIMIC-III. Visualisations of attention weights present a potential explainability tool for checking the face validity of ICD code predictions.
De-identifying Hospital Discharge Summaries: An End-to-End Framework using Ensemble of De-Identifiers
Jan 01, 2021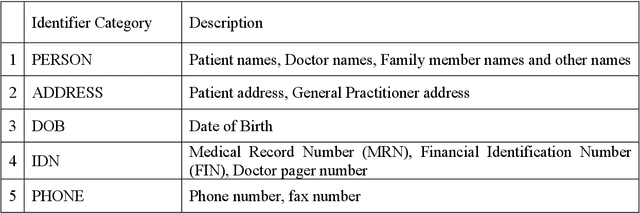
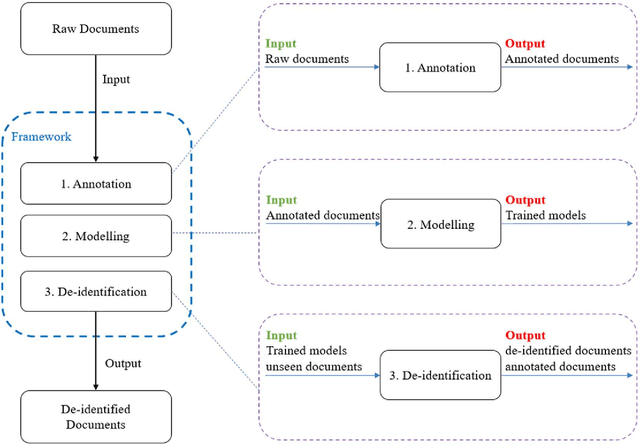
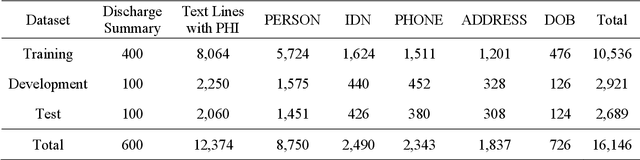
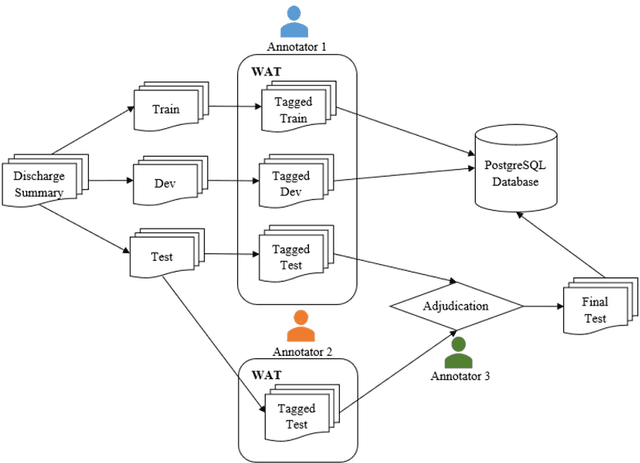
Abstract:Objective:Electronic Medical Records (EMRs) contain clinical narrative text that is of great potential value to medical researchers. However, this information is mixed with Protected Health Information (PHI) that presents risks to patient and clinician confidentiality. This paper presents an end-to-end de-identification framework to automatically remove PHI from hospital discharge summaries. Materials and Methods:Our corpus included 600 hospital discharge summaries which were extracted from the EMRs of two principal referral hospitals in Sydney, Australia. Our end-to-end de-identification framework consists of three components: 1) Annotation: labelling of PHI in the 600 hospital discharge summaries using five pre-defined categories: person, address, date of birth, individual identification number, phone/fax number; 2) Modelling: training and evaluating ensembles of named entity recognition (NER) models through the use of three natural language processing (NLP) toolkits (Stanza, FLAIR and spaCy) and both balanced and imbalanced datasets; and 3) De-identification: removing PHI from the hospital discharge summaries. Results:The final model in our framework was an ensemble which combined six single models using both balanced and imbalanced datasets for training majority voting. It achieved 0.9866 precision, 0.9862 recall and 0.9864 F1 scores. The majority of false positives and false negatives were related to the person category. Discussion:Our study showed that the ensemble of different models which were trained using three different NLP toolkits upon balanced and imbalanced datasets can achieve good results even with a relatively small corpus. Conclusion:Our end-to-end framework provides a robust solution to de-identifying clinical narrative corpuses safely. It can be easily applied to any kind of clinical narrative documents.
A Deep Representation of Longitudinal EMR Data Used for Predicting Readmission to the ICU and Describing Patients-at-Risk
May 21, 2019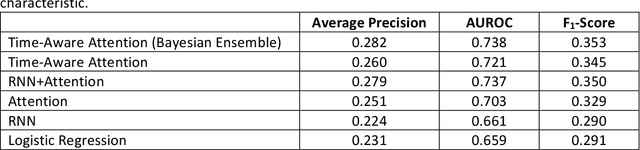
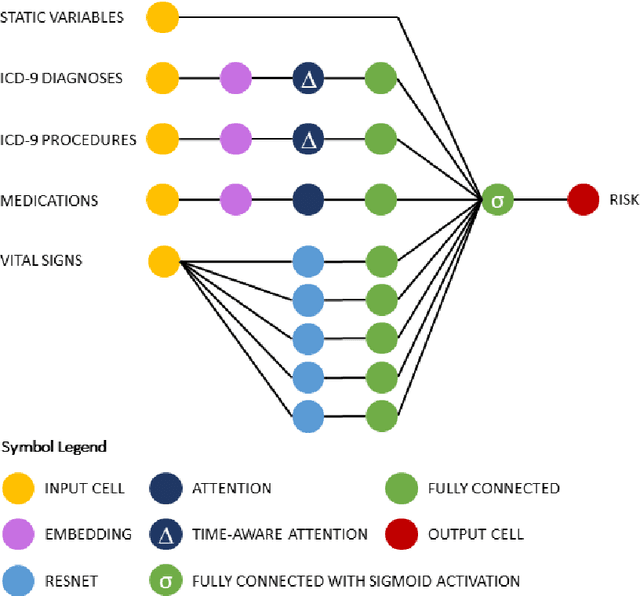
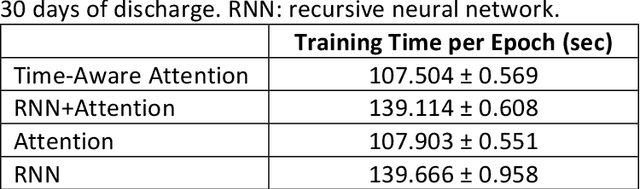
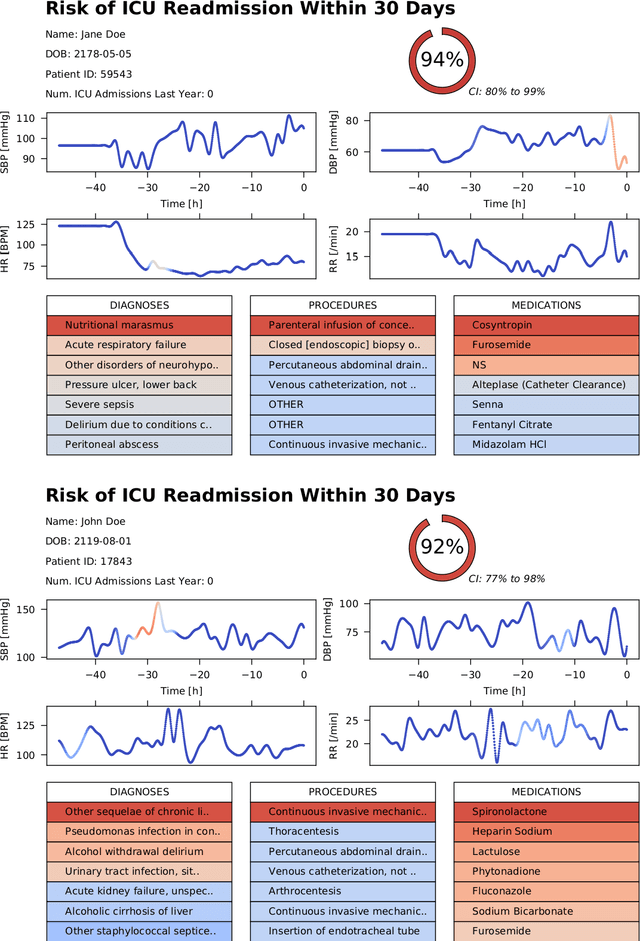
Abstract:Objective: To evaluate the feasibility of using an attention-based neural network for predicting the risk of readmission within 30 days of discharge from the intensive care unit (ICU) based on longitudinal electronic medical record (EMR) data and to leverage the interpretability of the model to describe patients-at-risk. Methods: A "time-aware attention" model was trained using publicly available EMR data (MIMIC-III) associated with 45,298 ICU stays for 33,150 patients. The analysed EMR data included static (patient demographics) and timestamped variables (diagnoses, procedures, medications, and vital signs). Bayesian inference was used to compute the posterior distribution of network weights. The prediction accuracy of the proposed model was compared with several baseline models and evaluated based on average precision, AUROC, and F1-Score. Odds ratios (ORs) associated with an increased risk of readmission were computed for static variables. Diagnoses, procedures, and medications were ranked according to the associated risk of readmission. The model was also used to generate reports with predicted risk (and associated uncertainty) justified by specific diagnoses, procedures, medications, and vital signs. Results: A Bayesian ensemble of 10 time-aware attention models led to the highest predictive accuracy (average precision: 0.282, AUROC: 0.738, F1-Score: 0.353). Male gender, number of recent admissions, age, admission location, insurance type, and ethnicity were all associated with risk of readmission. A longer length of stay in the ICU was found to reduce the risk of readmission (OR: 0.909, 95% credible interval: 0.902, 0.916). Groups of patients at risk included those requiring cardiovascular or ventilatory support, those with poor nutritional state, and those for whom standard medical care was not suitable, e.g. due to contraindications to surgery or medications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge