Alejandro Metke-Jimenez
Continuous time recurrent neural networks: overview and application to forecasting blood glucose in the intensive care unit
Apr 14, 2023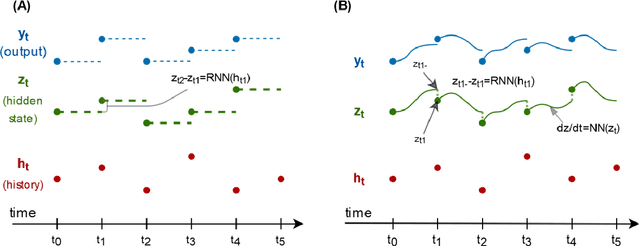
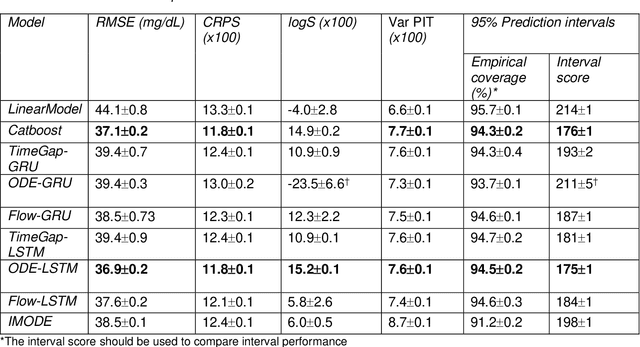
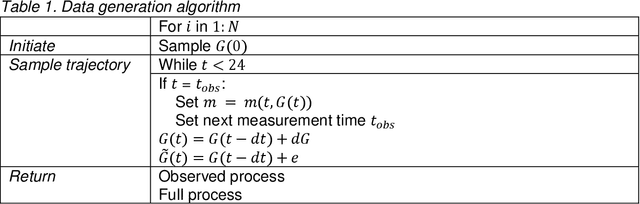
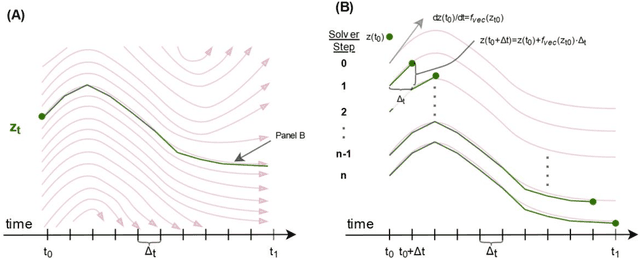
Abstract:Irregularly measured time series are common in many of the applied settings in which time series modelling is a key statistical tool, including medicine. This provides challenges in model choice, often necessitating imputation or similar strategies. Continuous time autoregressive recurrent neural networks (CTRNNs) are a deep learning model that account for irregular observations through incorporating continuous evolution of the hidden states between observations. This is achieved using a neural ordinary differential equation (ODE) or neural flow layer. In this manuscript, we give an overview of these models, including the varying architectures that have been proposed to account for issues such as ongoing medical interventions. Further, we demonstrate the application of these models to probabilistic forecasting of blood glucose in a critical care setting using electronic medical record and simulated data. The experiments confirm that addition of a neural ODE or neural flow layer generally improves the performance of autoregressive recurrent neural networks in the irregular measurement setting. However, several CTRNN architecture are outperformed by an autoregressive gradient boosted tree model (Catboost), with only a long short-term memory (LSTM) and neural ODE based architecture (ODE-LSTM) achieving comparable performance on probabilistic forecasting metrics such as the continuous ranked probability score (ODE-LSTM: 0.118$\pm$0.001; Catboost: 0.118$\pm$0.001), ignorance score (0.152$\pm$0.008; 0.149$\pm$0.002) and interval score (175$\pm$1; 176$\pm$1).
Semantic Search for Large Scale Clinical Ontologies
Jan 01, 2022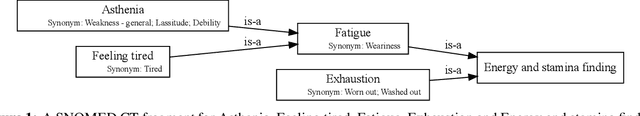
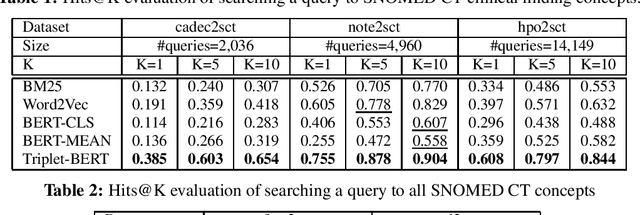
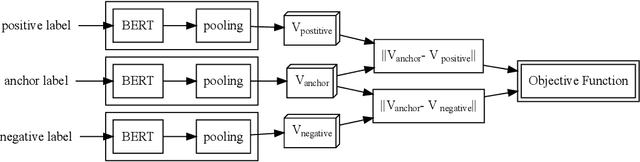
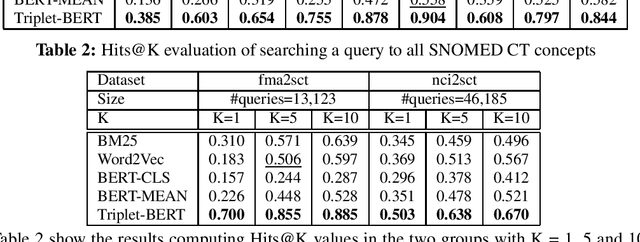
Abstract:Finding concepts in large clinical ontologies can be challenging when queries use different vocabularies. A search algorithm that overcomes this problem is useful in applications such as concept normalisation and ontology matching, where concepts can be referred to in different ways, using different synonyms. In this paper, we present a deep learning based approach to build a semantic search system for large clinical ontologies. We propose a Triplet-BERT model and a method that generates training data directly from the ontologies. The model is evaluated using five real benchmark data sets and the results show that our approach achieves high results on both free text to concept and concept to concept searching tasks, and outperforms all baseline methods.
Concept Extraction to Identify Adverse Drug Reactions in Medical Forums: A Comparison of Algorithms
Apr 27, 2015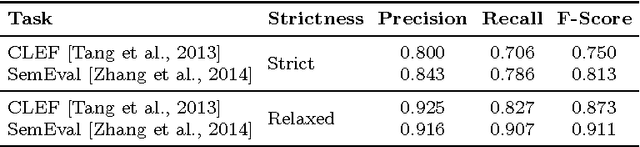
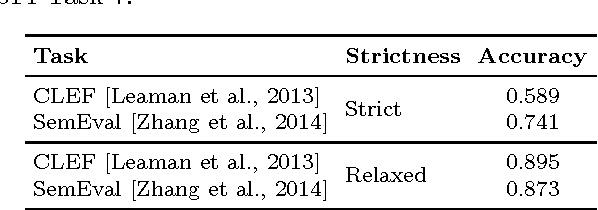
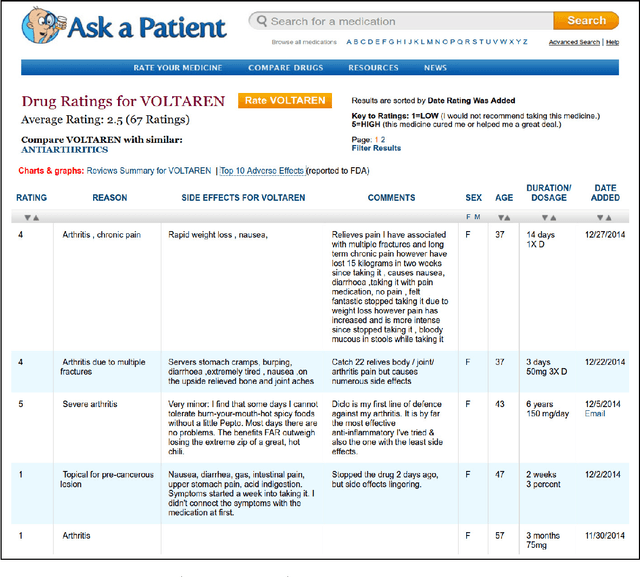
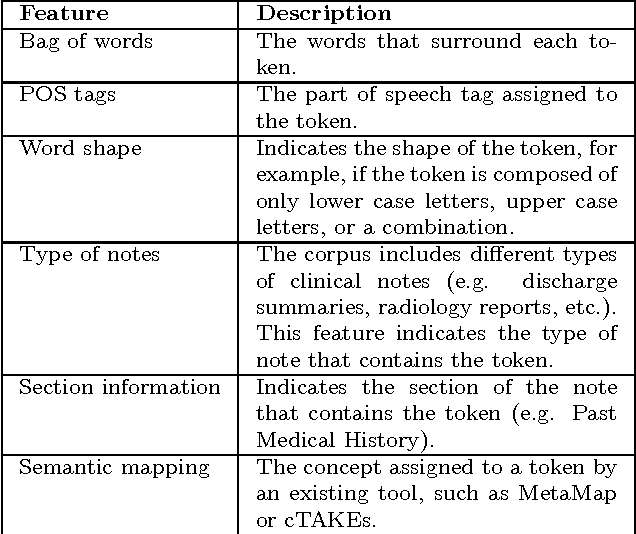
Abstract:Social media is becoming an increasingly important source of information to complement traditional pharmacovigilance methods. In order to identify signals of potential adverse drug reactions, it is necessary to first identify medical concepts in the social media text. Most of the existing studies use dictionary-based methods which are not evaluated independently from the overall signal detection task. We compare different approaches to automatically identify and normalise medical concepts in consumer reviews in medical forums. Specifically, we implement several dictionary-based methods popular in the relevant literature, as well as a method we suggest based on a state-of-the-art machine learning method for entity recognition. MetaMap, a popular biomedical concept extraction tool, is used as a baseline. Our evaluations were performed in a controlled setting on a common corpus which is a collection of medical forum posts annotated with concepts and linked to controlled vocabularies such as MedDRA and SNOMED CT. To our knowledge, our study is the first to systematically examine the effect of popular concept extraction methods in the area of signal detection for adverse reactions. We show that the choice of algorithm or controlled vocabulary has a significant impact on concept extraction, which will impact the overall signal detection process. We also show that our proposed machine learning approach significantly outperforms all the other methods in identification of both adverse reactions and drugs, even when trained with a relatively small set of annotated text.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge