Sarvnaz Karimi
Can AI Extract Antecedent Factors of Human Trust in AI? An Application of Information Extraction for Scientific Literature in Behavioural and Computer Sciences
Dec 16, 2024



Abstract:Information extraction from the scientific literature is one of the main techniques to transform unstructured knowledge hidden in the text into structured data which can then be used for decision-making in down-stream tasks. One such area is Trust in AI, where factors contributing to human trust in artificial intelligence applications are studied. The relationships of these factors with human trust in such applications are complex. We hence explore this space from the lens of information extraction where, with the input of domain experts, we carefully design annotation guidelines, create the first annotated English dataset in this domain, investigate an LLM-guided annotation, and benchmark it with state-of-the-art methods using large language models in named entity and relation extraction. Our results indicate that this problem requires supervised learning which may not be currently feasible with prompt-based LLMs.
AskBeacon -- Performing genomic data exchange and analytics with natural language
Oct 23, 2024



Abstract:Enabling clinicians and researchers to directly interact with global genomic data resources by removing technological barriers is vital for medical genomics. AskBeacon enables Large Language Models to be applied to securely shared cohorts via the GA4GH Beacon protocol. By simply "asking" Beacon, actionable insights can be gained, analyzed and made publication-ready.
A Critical Look at Meta-evaluating Summarisation Evaluation Metrics
Sep 29, 2024Abstract:Effective summarisation evaluation metrics enable researchers and practitioners to compare different summarisation systems efficiently. Estimating the effectiveness of an automatic evaluation metric, termed meta-evaluation, is a critically important research question. In this position paper, we review recent meta-evaluation practices for summarisation evaluation metrics and find that (1) evaluation metrics are primarily meta-evaluated on datasets consisting of examples from news summarisation datasets, and (2) there has been a noticeable shift in research focus towards evaluating the faithfulness of generated summaries. We argue that the time is ripe to build more diverse benchmarks that enable the development of more robust evaluation metrics and analyze the generalization ability of existing evaluation metrics. In addition, we call for research focusing on user-centric quality dimensions that consider the generated summary's communicative goal and the role of summarisation in the workflow.
MultiADE: A Multi-domain Benchmark for Adverse Drug Event Extraction
May 28, 2024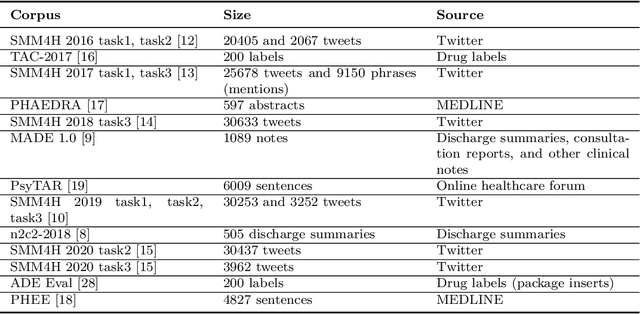
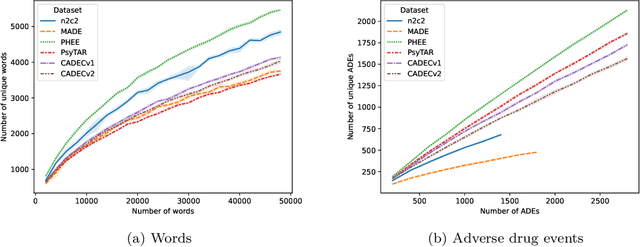
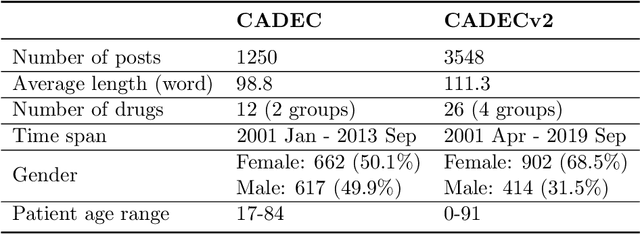
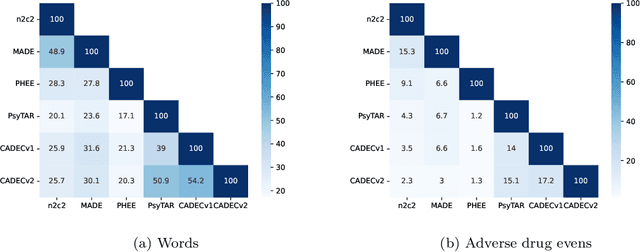
Abstract:Objective. Active adverse event surveillance monitors Adverse Drug Events (ADE) from different data sources, such as electronic health records, medical literature, social media and search engine logs. Over years, many datasets are created, and shared tasks are organised to facilitate active adverse event surveillance. However, most-if not all-datasets or shared tasks focus on extracting ADEs from a particular type of text. Domain generalisation-the ability of a machine learning model to perform well on new, unseen domains (text types)-is under-explored. Given the rapid advancements in natural language processing, one unanswered question is how far we are from having a single ADE extraction model that are effective on various types of text, such as scientific literature and social media posts}. Methods. We contribute to answering this question by building a multi-domain benchmark for adverse drug event extraction, which we named MultiADE. The new benchmark comprises several existing datasets sampled from different text types and our newly created dataset-CADECv2, which is an extension of CADEC (Karimi, et al., 2015), covering online posts regarding more diverse drugs than CADEC. Our new dataset is carefully annotated by human annotators following detailed annotation guidelines. Conclusion. Our benchmark results show that the generalisation of the trained models is far from perfect, making it infeasible to be deployed to process different types of text. In addition, although intermediate transfer learning is a promising approach to utilising existing resources, further investigation is needed on methods of domain adaptation, particularly cost-effective methods to select useful training instances.
Identifying Health Risks from Family History: A Survey of Natural Language Processing Techniques
Mar 15, 2024Abstract:Electronic health records include information on patients' status and medical history, which could cover the history of diseases and disorders that could be hereditary. One important use of family history information is in precision health, where the goal is to keep the population healthy with preventative measures. Natural Language Processing (NLP) and machine learning techniques can assist with identifying information that could assist health professionals in identifying health risks before a condition is developed in their later years, saving lives and reducing healthcare costs. We survey the literature on the techniques from the NLP field that have been developed to utilise digital health records to identify risks of familial diseases. We highlight that rule-based methods are heavily investigated and are still actively used for family history extraction. Still, more recent efforts have been put into building neural models based on large-scale pre-trained language models. In addition to the areas where NLP has successfully been utilised, we also identify the areas where more research is needed to unlock the value of patients' records regarding data collection, task formulation and downstream applications.
Detecting Entities in the Astrophysics Literature: A Comparison of Word-based and Span-based Entity Recognition Methods
Nov 24, 2022
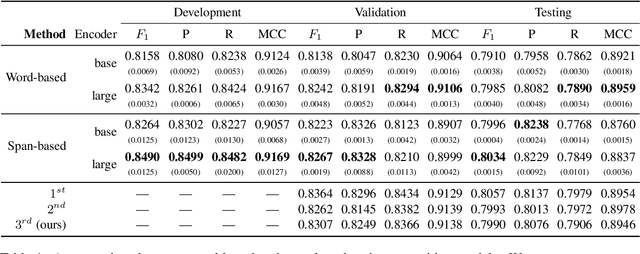
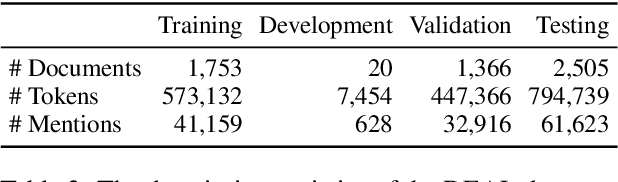

Abstract:Information Extraction from scientific literature can be challenging due to the highly specialised nature of such text. We describe our entity recognition methods developed as part of the DEAL (Detecting Entities in the Astrophysics Literature) shared task. The aim of the task is to build a system that can identify Named Entities in a dataset composed by scholarly articles from astrophysics literature. We planned our participation such that it enables us to conduct an empirical comparison between word-based tagging and span-based classification methods. When evaluated on two hidden test sets provided by the organizer, our best-performing submission achieved $F_1$ scores of 0.8307 (validation phase) and 0.7990 (testing phase).
Cost-effective Selection of Pretraining Data: A Case Study of Pretraining BERT on Social Media
Oct 02, 2020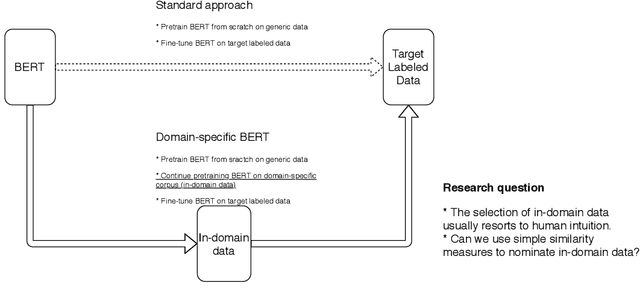
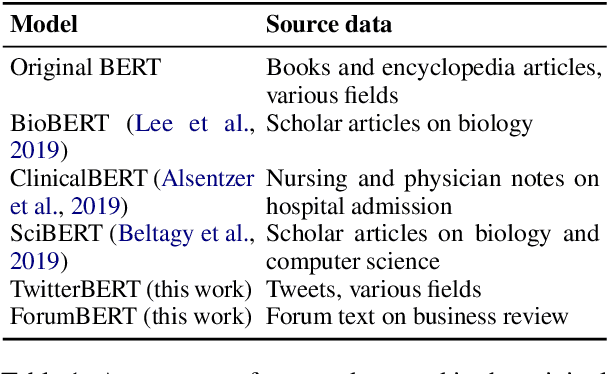
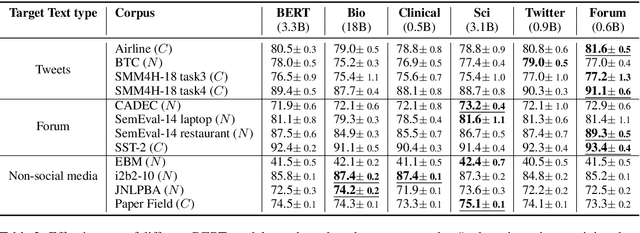
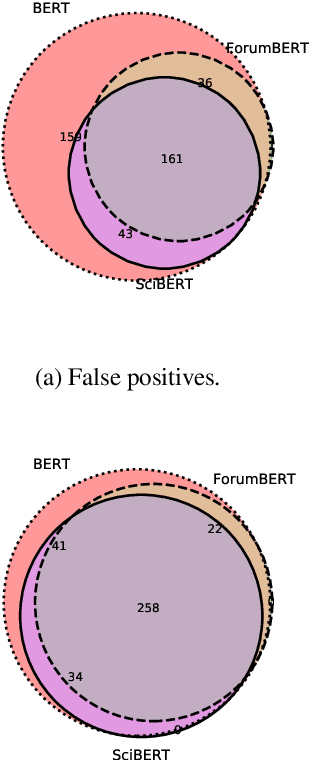
Abstract:Recent studies on domain-specific BERT models show that effectiveness on downstream tasks can be improved when models are pretrained on in-domain data. Often, the pretraining data used in these models are selected based on their subject matter, e.g., biology or computer science. Given the range of applications using social media text, and its unique language variety, we pretrain two models on tweets and forum text respectively, and empirically demonstrate the effectiveness of these two resources. In addition, we investigate how similarity measures can be used to nominate in-domain pretraining data. We publicly release our pretrained models at https://bit.ly/35RpTf0.
Searching Scientific Literature for Answers on COVID-19 Questions
Jul 06, 2020
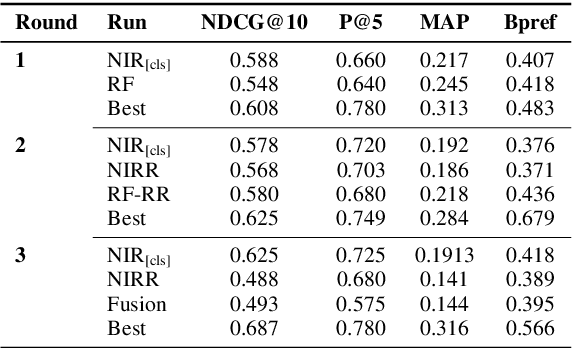
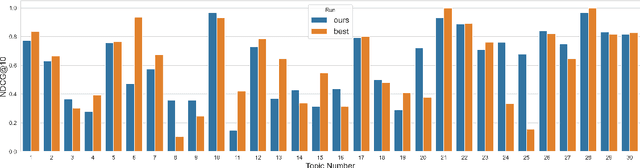
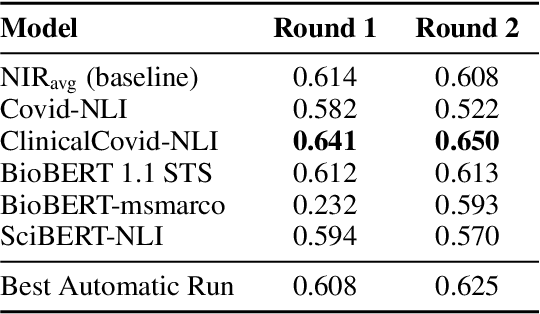
Abstract:Finding answers related to a pandemic of a novel disease raises new challenges for information seeking and retrieval, as the new information becomes available gradually. TREC COVID search track aims to assist in creating search tools to aid scientists, clinicians, policy makers and others with similar information needs in finding reliable answers from the scientific literature. We experiment with different ranking algorithms as part of our participation in this challenge. We propose a novel method for neural retrieval, and demonstrate its effectiveness on the TREC COVID search.
An Effective Transition-based Model for Discontinuous NER
Apr 28, 2020
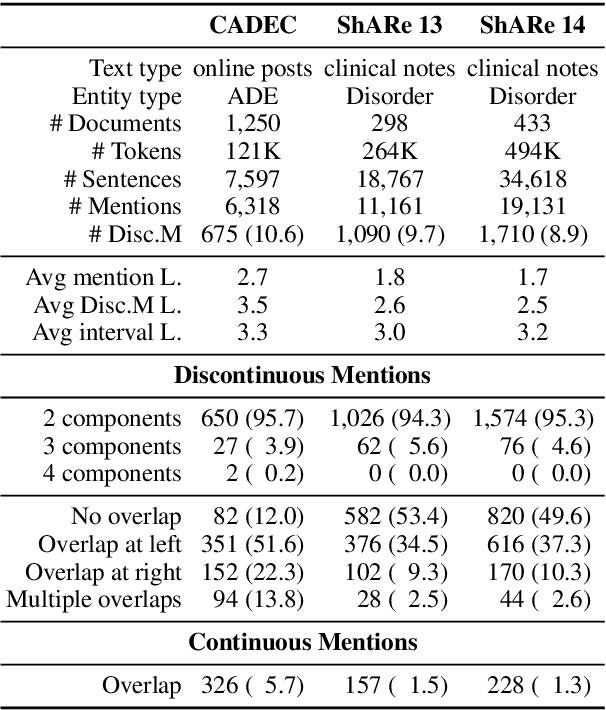
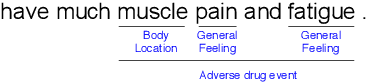
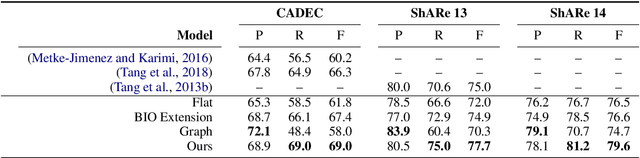
Abstract:Unlike widely used Named Entity Recognition (NER) data sets in generic domains, biomedical NER data sets often contain mentions consisting of discontinuous spans. Conventional sequence tagging techniques encode Markov assumptions that are efficient but preclude recovery of these mentions. We propose a simple, effective transition-based model with generic neural encoding for discontinuous NER. Through extensive experiments on three biomedical data sets, we show that our model can effectively recognize discontinuous mentions without sacrificing the accuracy on continuous mentions.
Figurative Usage Detection of Symptom Words to Improve Personal Health Mention Detection
Jul 04, 2019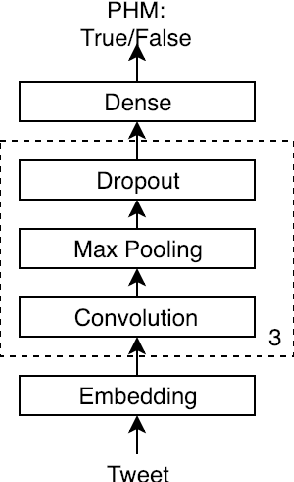
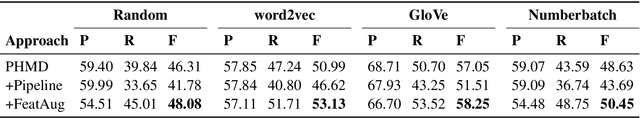
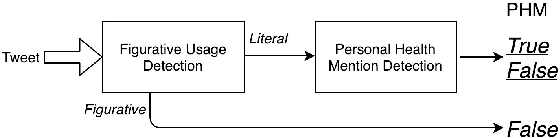
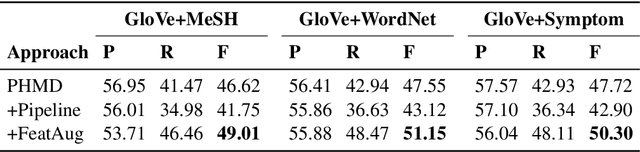
Abstract:Personal health mention detection deals with predicting whether or not a given sentence is a report of a health condition. Past work mentions errors in this prediction when symptom words, i.e. names of symptoms of interest, are used in a figurative sense. Therefore, we combine a state-of-the-art figurative usage detection with CNN-based personal health mention detection. To do so, we present two methods: a pipeline-based approach and a feature augmentation-based approach. The introduction of figurative usage detection results in an average improvement of 2.21% F-score of personal health mention detection, in the case of the feature augmentation-based approach. This paper demonstrates the promise of using figurative usage detection to improve personal health mention detection.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge