Sebastiano Barbieri
Queensland Digital Health Centre, University of Queensland, Brisbane, Australia, Centre for Big Data Research in Health, UNSW Sydney, Sydney, Australia
Generating Clinically Realistic EHR Data via a Hierarchy- and Semantics-Guided Transformer
Feb 28, 2025Abstract:Generating realistic synthetic electronic health records (EHRs) holds tremendous promise for accelerating healthcare research, facilitating AI model development and enhancing patient privacy. However, existing generative methods typically treat EHRs as flat sequences of discrete medical codes. This approach overlooks two critical aspects: the inherent hierarchical organization of clinical coding systems and the rich semantic context provided by code descriptions. Consequently, synthetic patient sequences often lack high clinical fidelity and have limited utility in downstream clinical tasks. In this paper, we propose the Hierarchy- and Semantics-Guided Transformer (HiSGT), a novel framework that leverages both hierarchical and semantic information for the generative process. HiSGT constructs a hierarchical graph to encode parent-child and sibling relationships among clinical codes and employs a graph neural network to derive hierarchy-aware embeddings. These are then fused with semantic embeddings extracted from a pre-trained clinical language model (e.g., ClinicalBERT), enabling the Transformer-based generator to more accurately model the nuanced clinical patterns inherent in real EHRs. Extensive experiments on the MIMIC-III and MIMIC-IV datasets demonstrate that HiSGT significantly improves the statistical alignment of synthetic data with real patient records, as well as supports robust downstream applications such as chronic disease classification. By addressing the limitations of conventional raw code-based generative models, HiSGT represents a significant step toward clinically high-fidelity synthetic data generation and a general paradigm suitable for interpretable medical code representation, offering valuable applications in data augmentation and privacy-preserving healthcare analytics.
Acquisition-Independent Deep Learning for Quantitative MRI Parameter Estimation using Neural Controlled Differential Equations
Dec 30, 2024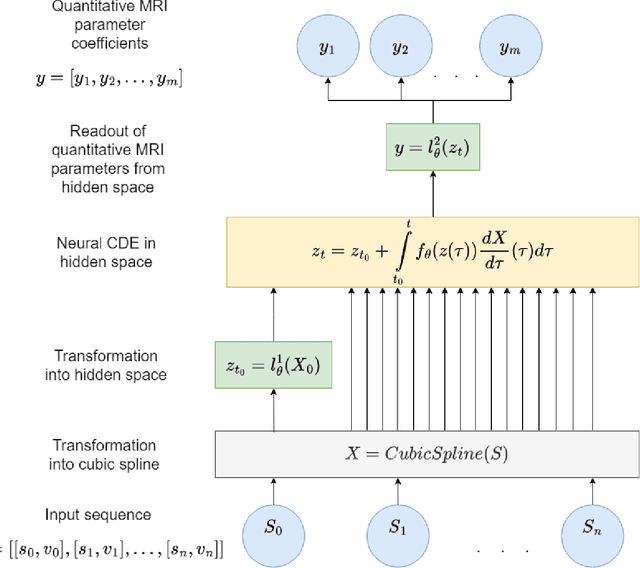
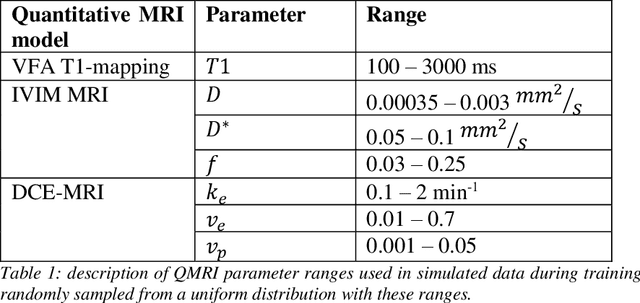
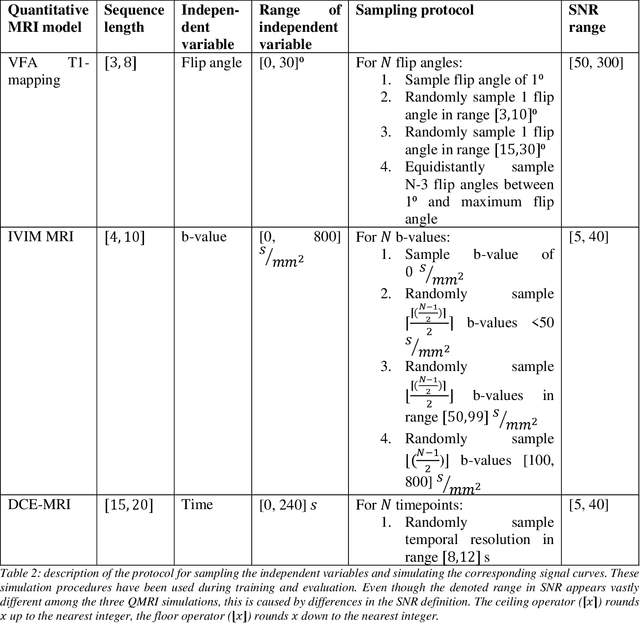
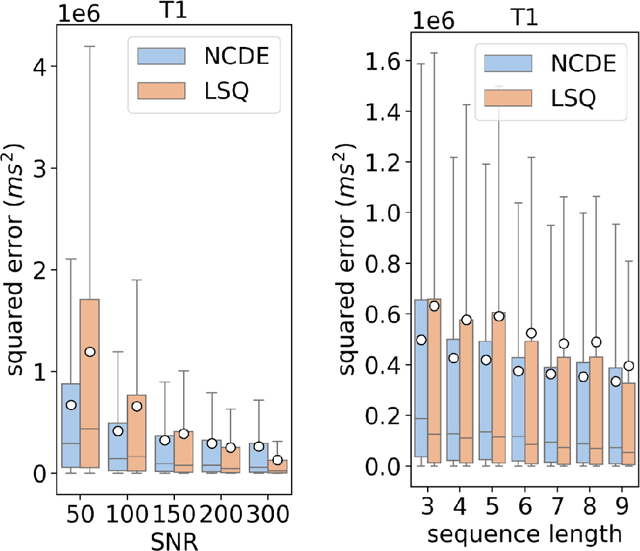
Abstract:Deep learning has proven to be a suitable alternative to least-squares (LSQ) fitting for parameter estimation in various quantitative MRI (QMRI) models. However, current deep learning implementations are not robust to changes in MR acquisition protocols. In practice, QMRI acquisition protocols differ substantially between different studies and clinical settings. The lack of generalizability and adoptability of current deep learning approaches for QMRI parameter estimation impedes the implementation of these algorithms in clinical trials and clinical practice. Neural Controlled Differential Equations (NCDEs) allow for the sampling of incomplete and irregularly sampled data with variable length, making them ideal for use in QMRI parameter estimation. In this study, we show that NCDEs can function as a generic tool for the accurate prediction of QMRI parameters, regardless of QMRI sequence length, configuration of independent variables and QMRI forward model (variable flip angle T1-mapping, intravoxel incoherent motion MRI, dynamic contrast-enhanced MRI). NCDEs achieved lower mean squared error than LSQ fitting in low-SNR simulations and in vivo in challenging anatomical regions like the abdomen and leg, but this improvement was no longer evident at high SNR. NCDEs reduce estimation error interquartile range without increasing bias, particularly under conditions of high uncertainty. These findings suggest that NCDEs offer a robust approach for reliable QMRI parameter estimation, especially in scenarios with high uncertainty or low image quality. We believe that with NCDEs, we have solved one of the main challenges for using deep learning for QMRI parameter estimation in a broader clinical and research setting.
Synthetic Health-related Longitudinal Data with Mixed-type Variables Generated using Diffusion Models
Mar 22, 2023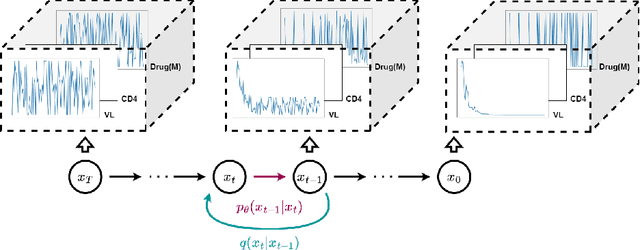

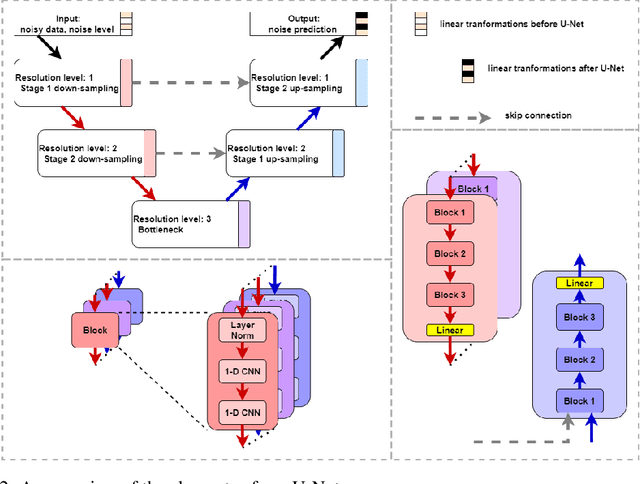
Abstract:This paper presents a novel approach to simulating electronic health records (EHRs) using diffusion probabilistic models (DPMs). Specifically, we demonstrate the effectiveness of DPMs in synthesising longitudinal EHRs that capture mixed-type variables, including numeric, binary, and categorical variables. To our knowledge, this represents the first use of DPMs for this purpose. We compared our DPM-simulated datasets to previous state-of-the-art results based on generative adversarial networks (GANs) for two clinical applications: acute hypotension and human immunodeficiency virus (ART for HIV). Given the lack of similar previous studies in DPMs, a core component of our work involves exploring the advantages and caveats of employing DPMs across a wide range of aspects. In addition to assessing the realism of the synthetic datasets, we also trained reinforcement learning (RL) agents on the synthetic data to evaluate their utility for supporting the development of downstream machine learning models. Finally, we estimated that our DPM-simulated datasets are secure and posed a low patient exposure risk for public access.
Generating Synthetic Clinical Data that Capture Class Imbalanced Distributions with Generative Adversarial Networks: Example using Antiretroviral Therapy for HIV
Aug 18, 2022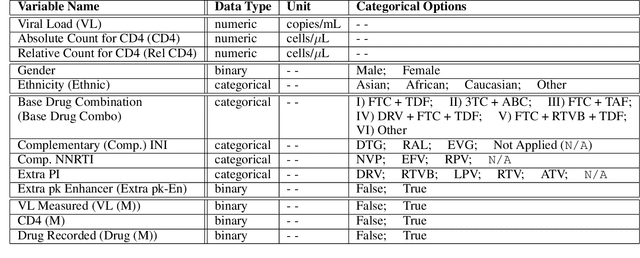
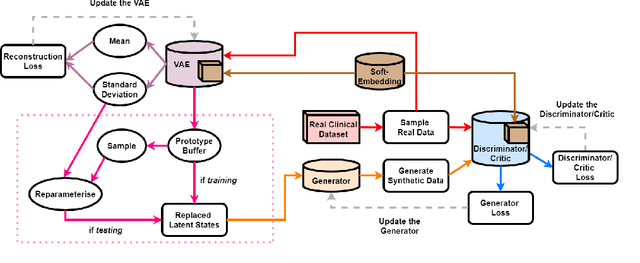
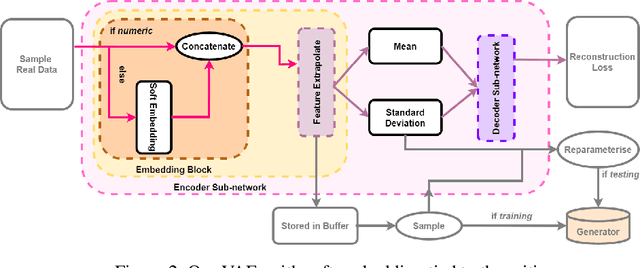
Abstract:Clinical data usually cannot be freely distributed due to their highly confidential nature and this hampers the development of machine learning in the healthcare domain. One way to mitigate this problem is by generating realistic synthetic datasets using generative adversarial networks (GANs). However, GANs are known to suffer from mode collapse and thus creating outputs of low diveristy. In this paper, we extend the classic GAN setup with an external memory to replay features from real samples. Using antiretroviral therapy for human immunodeficiency virus (ART for HIV) as a case study, we show that our extended setup increases convergence and more importantly, it is effective in capturing the severe class imbalanced distributions common to real world clinical data.
The Health Gym: Synthetic Health-Related Datasets for the Development of Reinforcement Learning Algorithms
Mar 12, 2022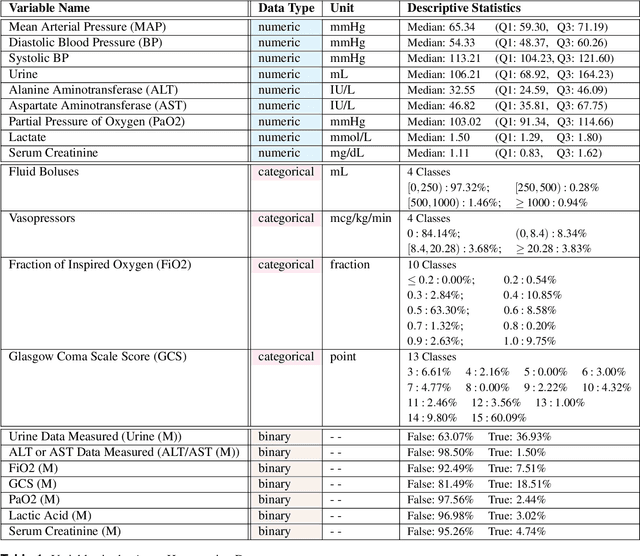
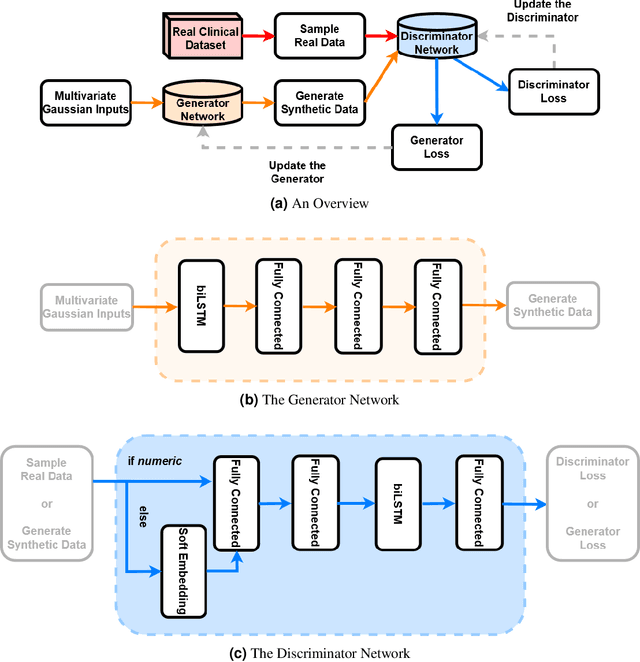
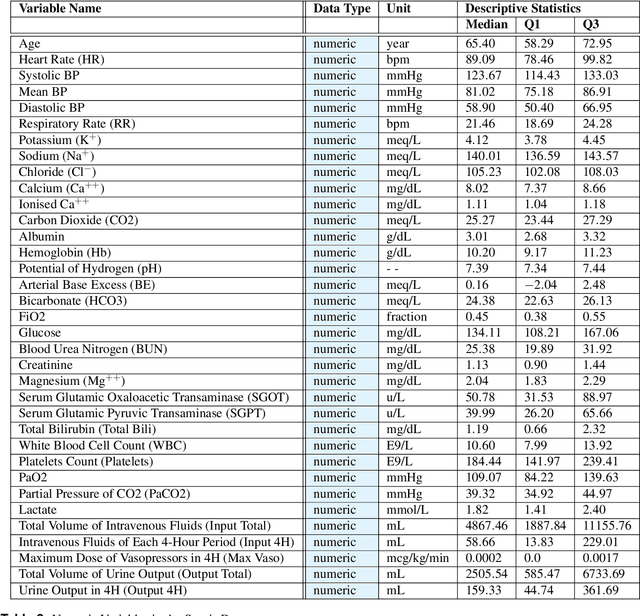
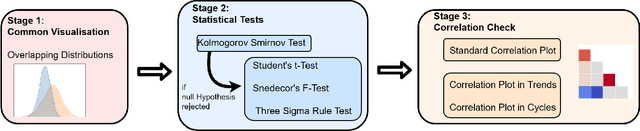
Abstract:In recent years, the machine learning research community has benefited tremendously from the availability of openly accessible benchmark datasets. Clinical data are usually not openly available due to their highly confidential nature. This has hampered the development of reproducible and generalisable machine learning applications in health care. Here we introduce the Health Gym - a growing collection of highly realistic synthetic medical datasets that can be freely accessed to prototype, evaluate, and compare machine learning algorithms, with a specific focus on reinforcement learning. The three synthetic datasets described in this paper present patient cohorts with acute hypotension and sepsis in the intensive care unit, and people with human immunodeficiency virus (HIV) receiving antiretroviral therapy in ambulatory care. The datasets were created using a novel generative adversarial network (GAN). The distributions of variables, and correlations between variables and trends over time in the synthetic datasets mirror those in the real datasets. Furthermore, the risk of sensitive information disclosure associated with the public distribution of the synthetic datasets is estimated to be very low.
Synthetic Acute Hypotension and Sepsis Datasets Based on MIMIC-III and Published as Part of the Health Gym Project
Dec 07, 2021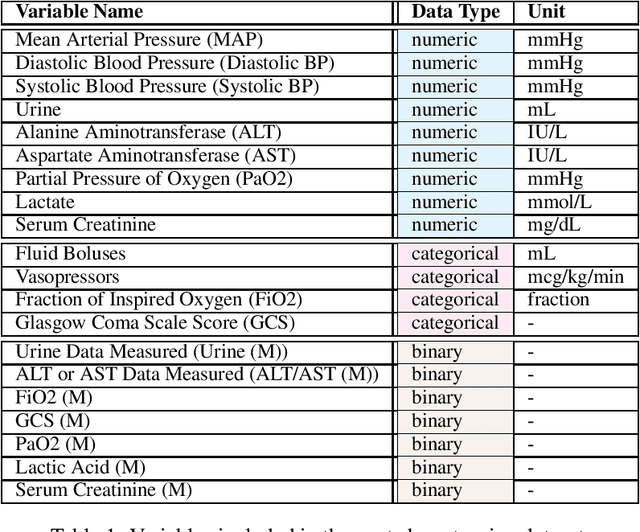

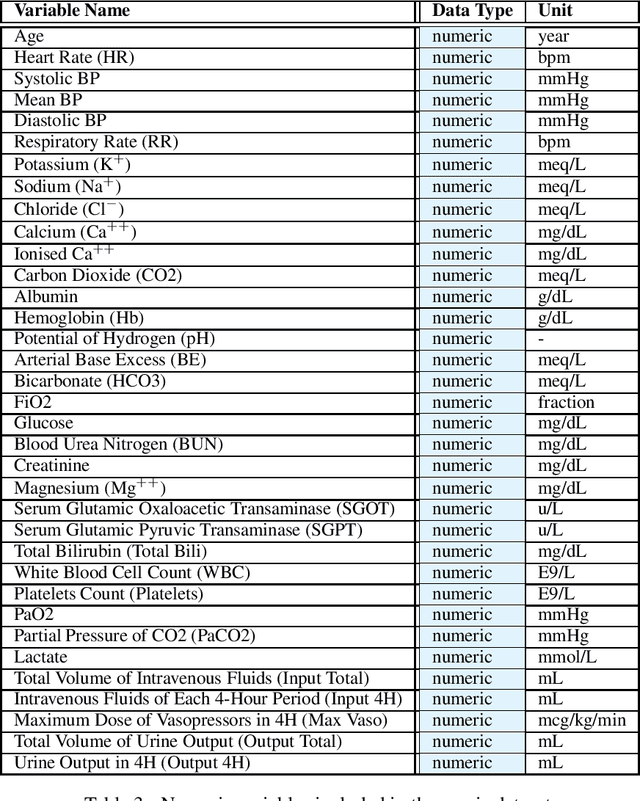
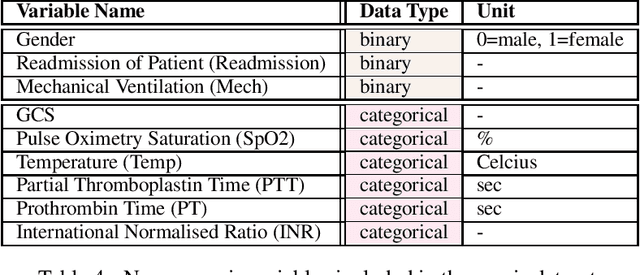
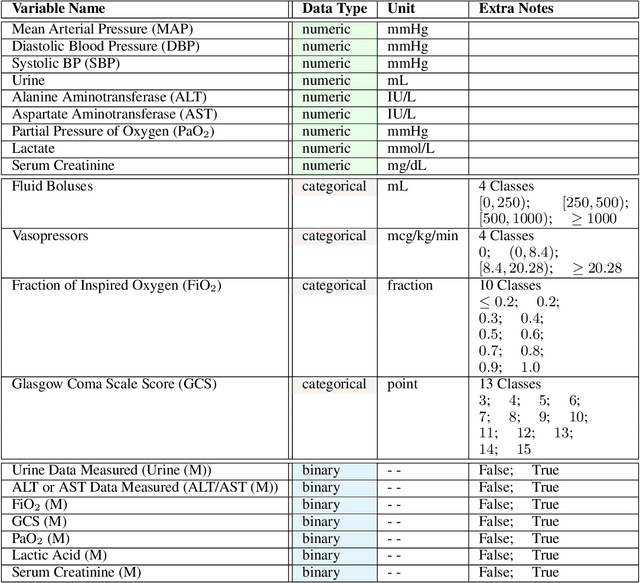
Abstract:These two synthetic datasets comprise vital signs, laboratory test results, administered fluid boluses and vasopressors for 3,910 patients with acute hypotension and for 2,164 patients with sepsis in the Intensive Care Unit (ICU). The patient cohorts were built using previously published inclusion and exclusion criteria and the data were created using Generative Adversarial Networks (GANs) and the MIMIC-III Clinical Database. The risk of identity disclosure associated with the release of these data was estimated to be very low (0.045%). The datasets were generated and published as part of the Health Gym, a project aiming to publicly distribute synthetic longitudinal health data for developing machine learning algorithms (with a particular focus on offline reinforcement learning) and for educational purposes.
Incorporating Uncertainty in Learning to Defer Algorithms for Safe Computer-Aided Diagnosis
Sep 03, 2021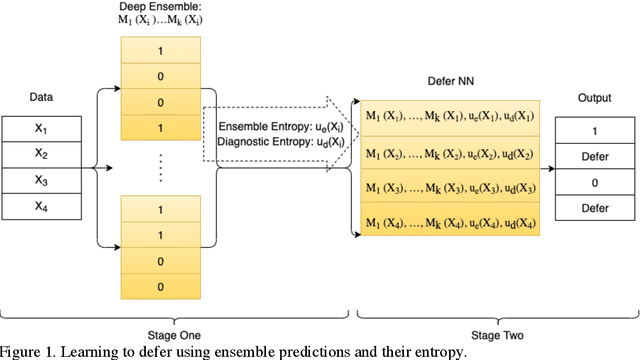
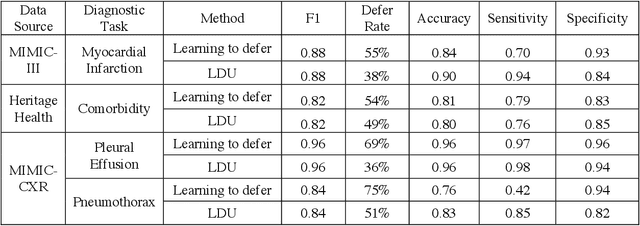
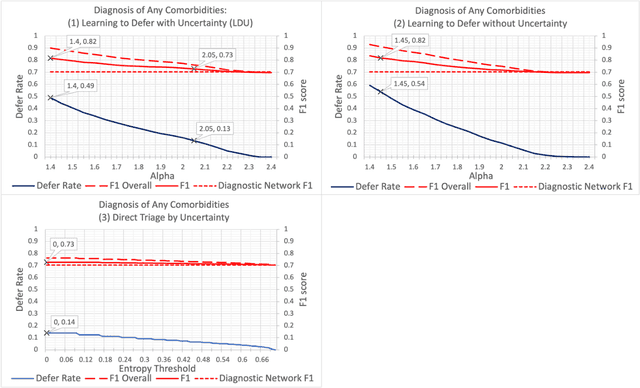
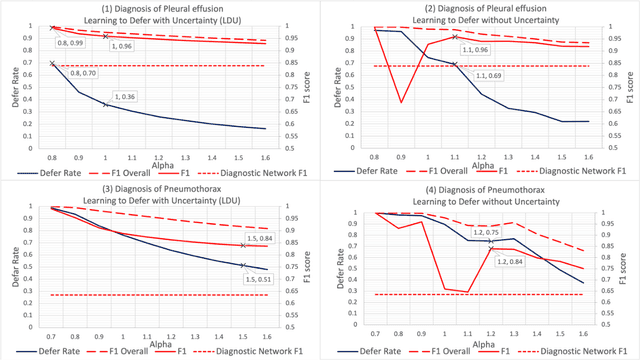
Abstract:In this study we propose the Learning to Defer with Uncertainty (LDU) algorithm, an approach which considers the model's predictive uncertainty when identifying the patient group to be evaluated by human experts. By identifying patients for whom the uncertainty of computer-aided diagnosis is estimated to be high and defers them for evaluation by human experts, the LDU algorithm can be used to mitigate the risk of erroneous computer-aided diagnoses in clinical settings.
Predicting cardiovascular risk from national administrative databases using a combined survival analysis and deep learning approach
Nov 28, 2020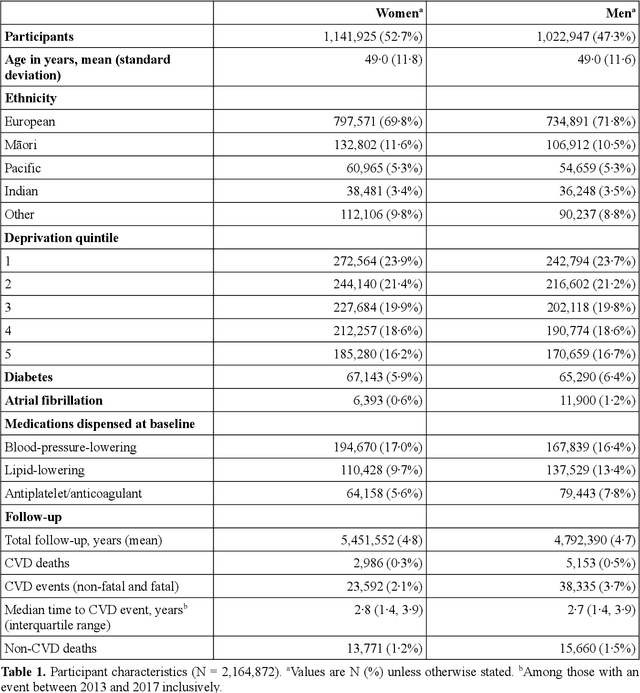
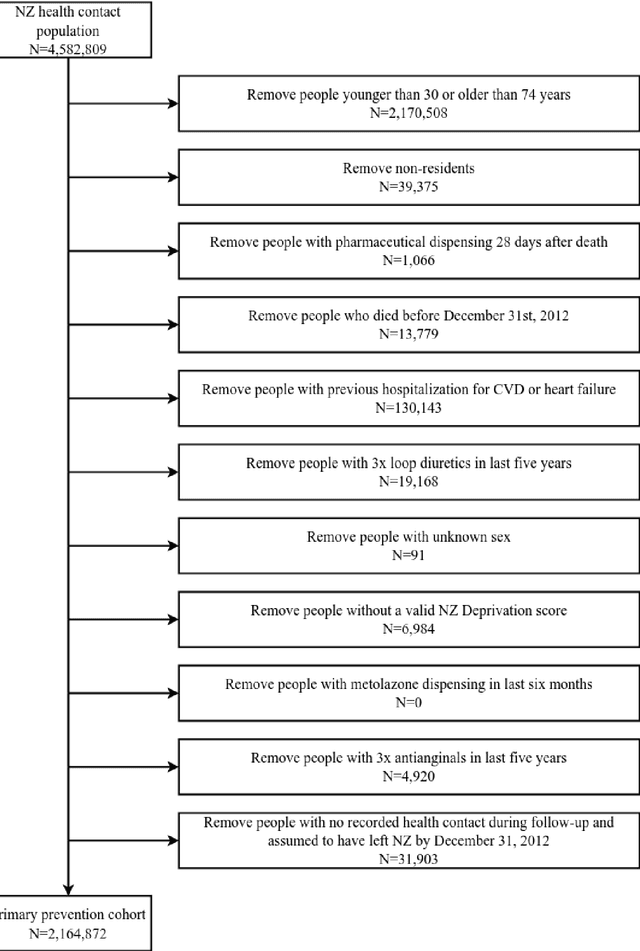
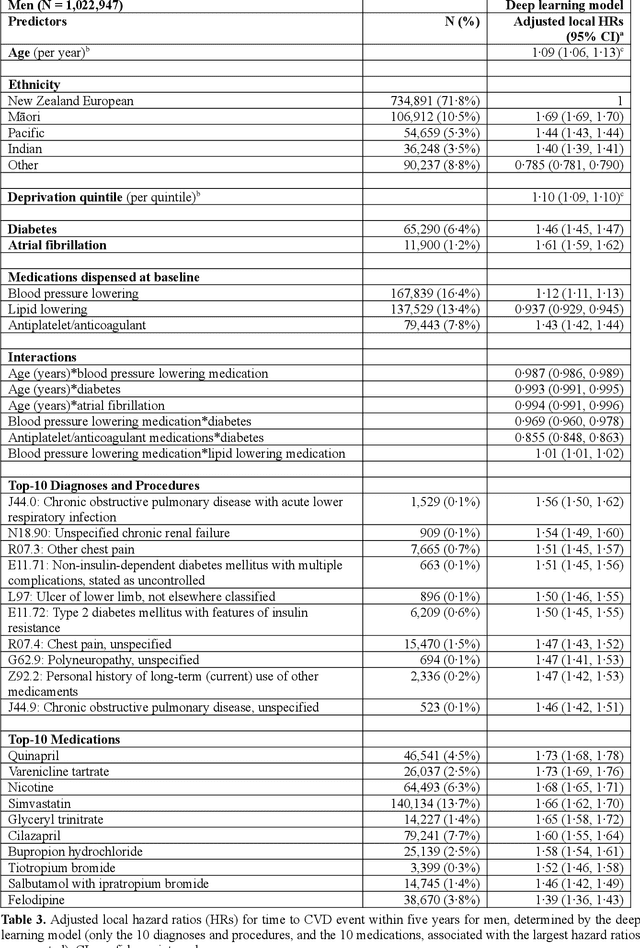
Abstract:AIMS. This study compared the performance of deep learning extensions of survival analysis models with traditional Cox proportional hazards (CPH) models for deriving cardiovascular disease (CVD) risk prediction equations in national health administrative datasets. METHODS. Using individual person linkage of multiple administrative datasets, we constructed a cohort of all New Zealand residents aged 30-74 years who interacted with publicly funded health services during 2012, and identified hospitalisations and deaths from CVD over five years of follow-up. After excluding people with prior CVD or heart failure, sex-specific deep learning and CPH models were developed to estimate the risk of fatal or non-fatal CVD events within five years. The proportion of explained time-to-event occurrence, calibration, and discrimination were compared between models across the whole study population and in specific risk groups. FINDINGS. First CVD events occurred in 61,927 of 2,164,872 people. Among diagnoses and procedures, the largest 'local' hazard ratios were associated by the deep learning models with tobacco use in women (2.04, 95%CI: 1.99-2.10) and with chronic obstructive pulmonary disease with acute lower respiratory infection in men (1.56, 95%CI: 1.50-1.62). Other identified predictors (e.g. hypertension, chest pain, diabetes) aligned with current knowledge about CVD risk predictors. The deep learning models significantly outperformed the CPH models on the basis of proportion of explained time-to-event occurrence (Royston and Sauerbrei's R-squared: 0.468 vs. 0.425 in women and 0.383 vs. 0.348 in men), calibration, and discrimination (all p<0.0001). INTERPRETATION. Deep learning extensions of survival analysis models can be applied to large health administrative databases to derive interpretable CVD risk prediction equations that are more accurate than traditional CPH models.
Improved unsupervised physics-informed deep learning for intravoxel-incoherent motion modeling and evaluation in pancreatic cancer patients
Nov 03, 2020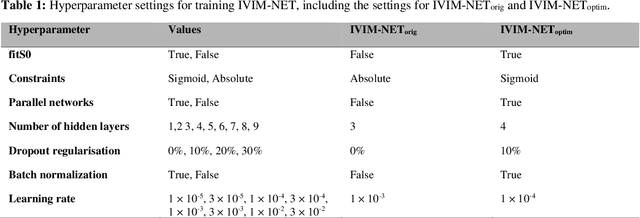
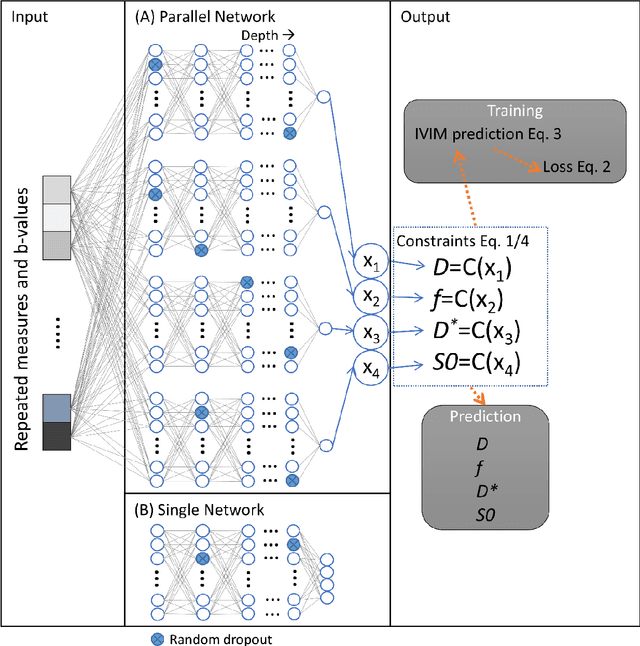
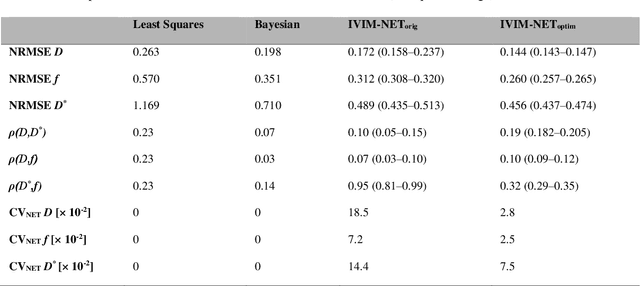
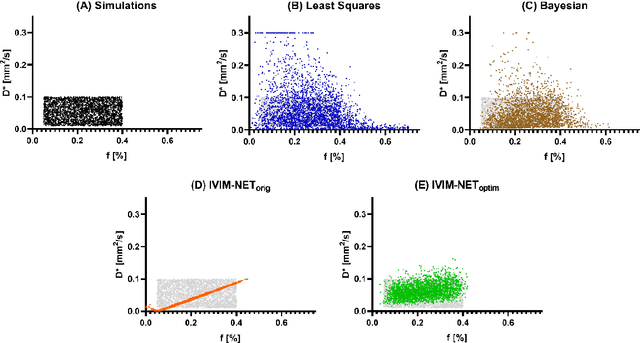
Abstract:${\bf Purpose}$: Earlier work showed that IVIM-NET$_{orig}$, an unsupervised physics-informed deep neural network, was faster and more accurate than other state-of-the-art intravoxel-incoherent motion (IVIM) fitting approaches to DWI. This study presents: IVIM-NET$_{optim}$, overcoming IVIM-NET$_{orig}$'s shortcomings. ${\bf Method}$: In simulations (SNR=20), the accuracy, independence and consistency of IVIM-NET were evaluated for combinations of hyperparameters (fit S0, constraints, network architecture, # hidden layers, dropout, batch normalization, learning rate), by calculating the NRMSE, Spearman's $\rho$, and the coefficient of variation (CV$_{NET}$), respectively. The best performing network, IVIM-NET$_{optim}$ was compared to least squares (LS) and a Bayesian approach at different SNRs. IVIM-NET$_{optim}$'s performance was evaluated in 23 pancreatic ductal adenocarcinoma (PDAC) patients. 14 of the patients received no treatment between 2 repeated scan sessions and 9 received chemoradiotherapy between sessions. Intersession within-subject standard deviations (wSD) and treatment-induced changes were assessed. ${\bf Results}$: In simulations, IVIM-NET$_{optim}$ outperformed IVIM-NET$_{orig}$ in accuracy (NRMSE(D)=0.14 vs 0.17; NMRSE(f)=0.26 vs 0.31; NMRSE(D*)=0.46 vs 0.49), independence ($\rho$(D*,f)=0.32 vs 0.95) and consistency (CV$_{NET}$ (D)=0.028 vs 0.185; CV$_{NET}$ (f)=0.025 vs 0.078; CV$_{NET}$ (D*)=0.075 vs 0.144). IVIM-NET$_{optim}$ showed superior performance to the LS and Bayesian approaches at SNRs<50. In vivo, IVIM-NET$_{optim}$ showed less noisy and more detailed parameter maps with lower wSD for D and f than the alternatives. In the treated cohort, IVIM-NET$_{optim}$ detected the most individual patients with significant parameter changes compared to day-to-day variations. ${\bf Conclusion}$: IVIM-NET$_{optim}$ is recommended for accurate IVIM fitting to DWI data.
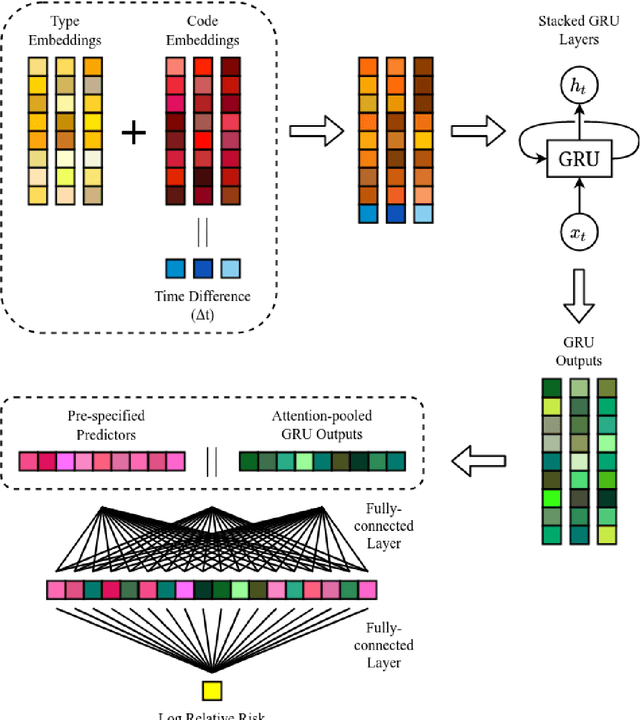
A Deep Representation of Longitudinal EMR Data Used for Predicting Readmission to the ICU and Describing Patients-at-Risk
May 21, 2019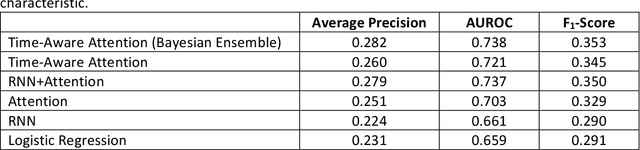
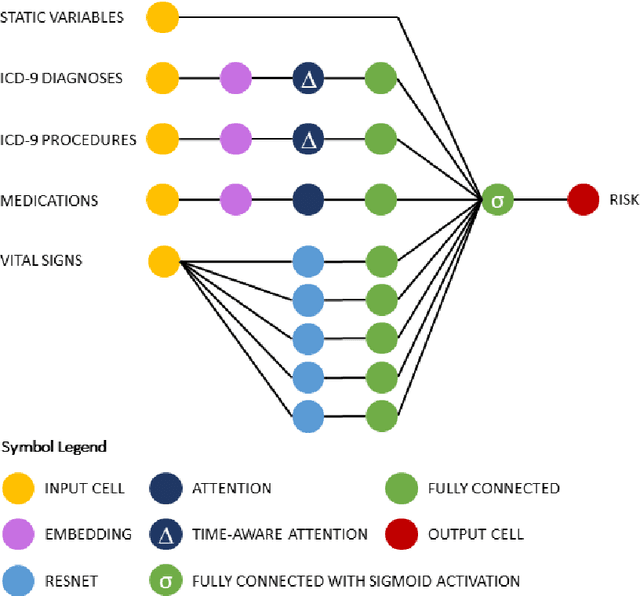
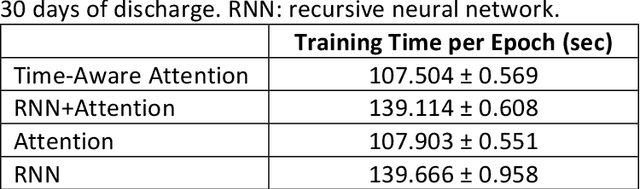
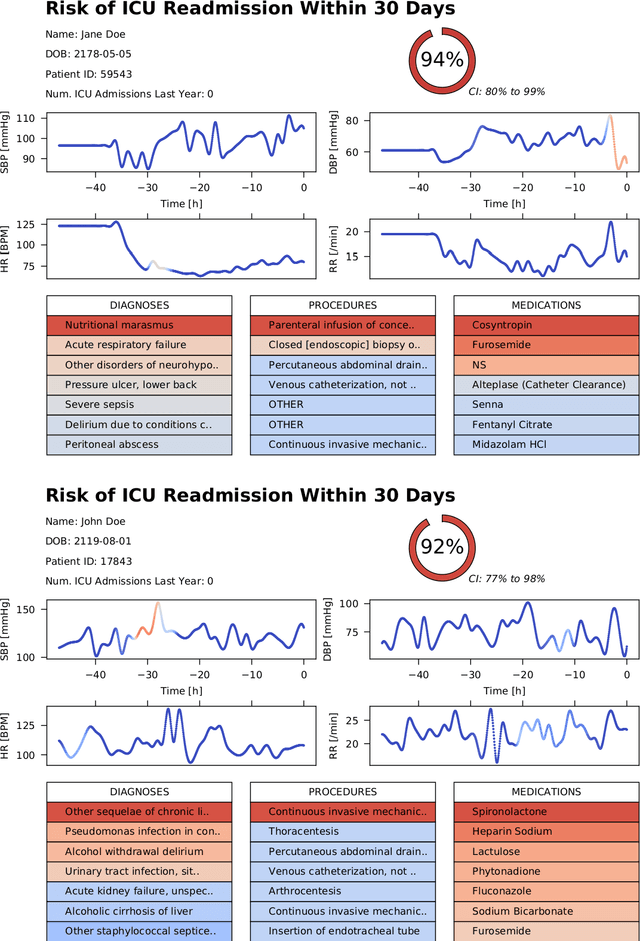
Abstract:Objective: To evaluate the feasibility of using an attention-based neural network for predicting the risk of readmission within 30 days of discharge from the intensive care unit (ICU) based on longitudinal electronic medical record (EMR) data and to leverage the interpretability of the model to describe patients-at-risk. Methods: A "time-aware attention" model was trained using publicly available EMR data (MIMIC-III) associated with 45,298 ICU stays for 33,150 patients. The analysed EMR data included static (patient demographics) and timestamped variables (diagnoses, procedures, medications, and vital signs). Bayesian inference was used to compute the posterior distribution of network weights. The prediction accuracy of the proposed model was compared with several baseline models and evaluated based on average precision, AUROC, and F1-Score. Odds ratios (ORs) associated with an increased risk of readmission were computed for static variables. Diagnoses, procedures, and medications were ranked according to the associated risk of readmission. The model was also used to generate reports with predicted risk (and associated uncertainty) justified by specific diagnoses, procedures, medications, and vital signs. Results: A Bayesian ensemble of 10 time-aware attention models led to the highest predictive accuracy (average precision: 0.282, AUROC: 0.738, F1-Score: 0.353). Male gender, number of recent admissions, age, admission location, insurance type, and ethnicity were all associated with risk of readmission. A longer length of stay in the ICU was found to reduce the risk of readmission (OR: 0.909, 95% credible interval: 0.902, 0.916). Groups of patients at risk included those requiring cardiovascular or ventilatory support, those with poor nutritional state, and those for whom standard medical care was not suitable, e.g. due to contraindications to surgery or medications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge