Oliver J. Gurney-Champion
Department of Radiology and Nuclear Medicine, Amsterdam University Medical Center, Amsterdam, The Netherlands, Imaging and Biomarkers, Cancer Center Amsterdam, Amsterdam, The Netherlands
Acquisition-Independent Deep Learning for Quantitative MRI Parameter Estimation using Neural Controlled Differential Equations
Dec 30, 2024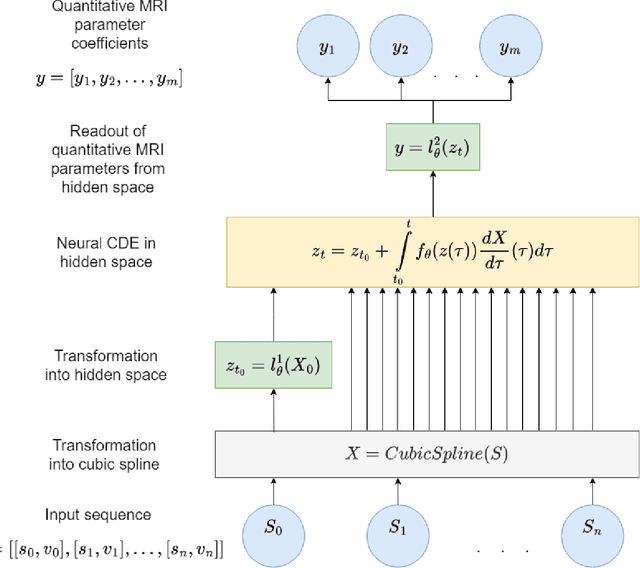
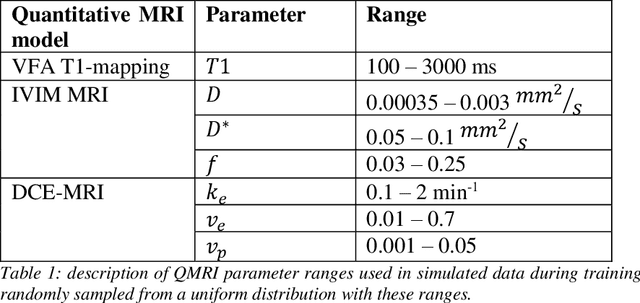
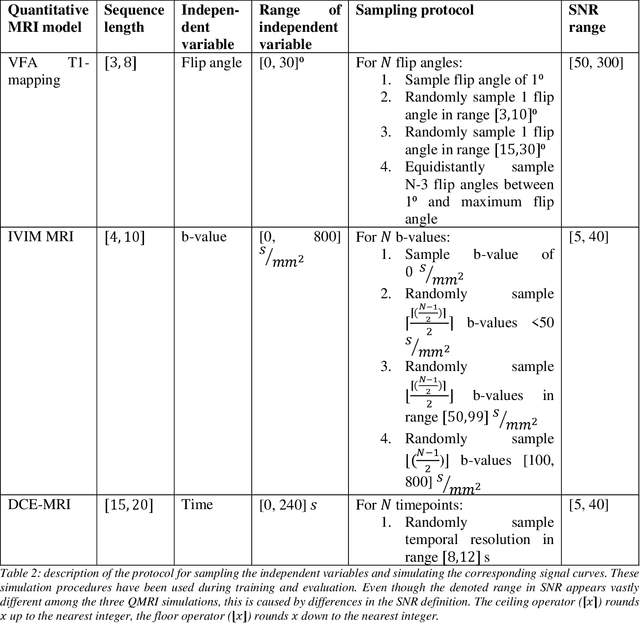
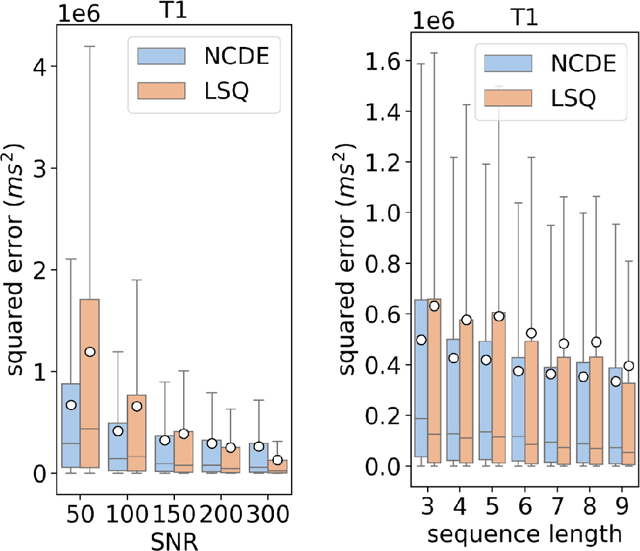
Abstract:Deep learning has proven to be a suitable alternative to least-squares (LSQ) fitting for parameter estimation in various quantitative MRI (QMRI) models. However, current deep learning implementations are not robust to changes in MR acquisition protocols. In practice, QMRI acquisition protocols differ substantially between different studies and clinical settings. The lack of generalizability and adoptability of current deep learning approaches for QMRI parameter estimation impedes the implementation of these algorithms in clinical trials and clinical practice. Neural Controlled Differential Equations (NCDEs) allow for the sampling of incomplete and irregularly sampled data with variable length, making them ideal for use in QMRI parameter estimation. In this study, we show that NCDEs can function as a generic tool for the accurate prediction of QMRI parameters, regardless of QMRI sequence length, configuration of independent variables and QMRI forward model (variable flip angle T1-mapping, intravoxel incoherent motion MRI, dynamic contrast-enhanced MRI). NCDEs achieved lower mean squared error than LSQ fitting in low-SNR simulations and in vivo in challenging anatomical regions like the abdomen and leg, but this improvement was no longer evident at high SNR. NCDEs reduce estimation error interquartile range without increasing bias, particularly under conditions of high uncertainty. These findings suggest that NCDEs offer a robust approach for reliable QMRI parameter estimation, especially in scenarios with high uncertainty or low image quality. We believe that with NCDEs, we have solved one of the main challenges for using deep learning for QMRI parameter estimation in a broader clinical and research setting.
Improved unsupervised physics-informed deep learning for intravoxel-incoherent motion modeling and evaluation in pancreatic cancer patients
Nov 03, 2020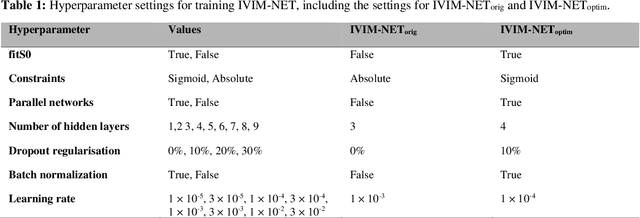
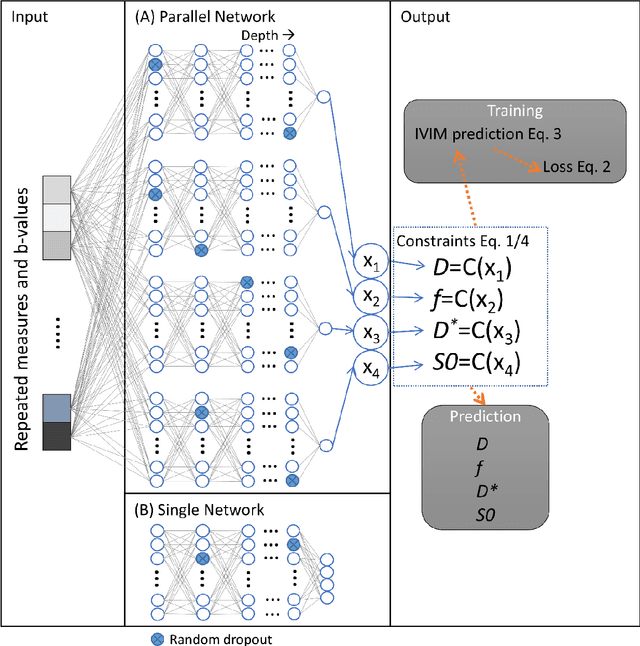
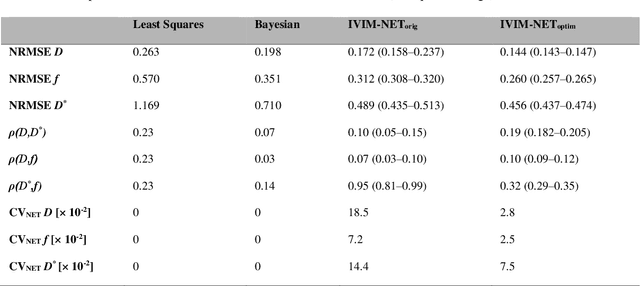
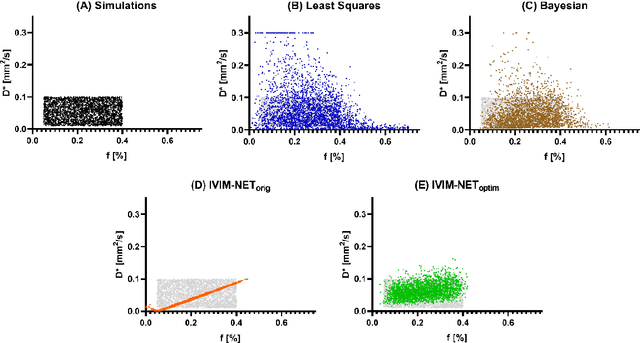
Abstract:${\bf Purpose}$: Earlier work showed that IVIM-NET$_{orig}$, an unsupervised physics-informed deep neural network, was faster and more accurate than other state-of-the-art intravoxel-incoherent motion (IVIM) fitting approaches to DWI. This study presents: IVIM-NET$_{optim}$, overcoming IVIM-NET$_{orig}$'s shortcomings. ${\bf Method}$: In simulations (SNR=20), the accuracy, independence and consistency of IVIM-NET were evaluated for combinations of hyperparameters (fit S0, constraints, network architecture, # hidden layers, dropout, batch normalization, learning rate), by calculating the NRMSE, Spearman's $\rho$, and the coefficient of variation (CV$_{NET}$), respectively. The best performing network, IVIM-NET$_{optim}$ was compared to least squares (LS) and a Bayesian approach at different SNRs. IVIM-NET$_{optim}$'s performance was evaluated in 23 pancreatic ductal adenocarcinoma (PDAC) patients. 14 of the patients received no treatment between 2 repeated scan sessions and 9 received chemoradiotherapy between sessions. Intersession within-subject standard deviations (wSD) and treatment-induced changes were assessed. ${\bf Results}$: In simulations, IVIM-NET$_{optim}$ outperformed IVIM-NET$_{orig}$ in accuracy (NRMSE(D)=0.14 vs 0.17; NMRSE(f)=0.26 vs 0.31; NMRSE(D*)=0.46 vs 0.49), independence ($\rho$(D*,f)=0.32 vs 0.95) and consistency (CV$_{NET}$ (D)=0.028 vs 0.185; CV$_{NET}$ (f)=0.025 vs 0.078; CV$_{NET}$ (D*)=0.075 vs 0.144). IVIM-NET$_{optim}$ showed superior performance to the LS and Bayesian approaches at SNRs<50. In vivo, IVIM-NET$_{optim}$ showed less noisy and more detailed parameter maps with lower wSD for D and f than the alternatives. In the treated cohort, IVIM-NET$_{optim}$ detected the most individual patients with significant parameter changes compared to day-to-day variations. ${\bf Conclusion}$: IVIM-NET$_{optim}$ is recommended for accurate IVIM fitting to DWI data.
Deep Learning How to Fit an Intravoxel Incoherent Motion Model to Diffusion-Weighted MRI
Feb 28, 2019



Abstract:Purpose: This prospective clinical study assesses the feasibility of training a deep neural network (DNN) for intravoxel incoherent motion (IVIM) model fitting to diffusion-weighted magnetic resonance imaging (DW-MRI) data and evaluates its performance. Methods: Approval for this study was obtained by the responsible ethics committees and written informed consent was obtained from all accrued subjects. In May 2011, ten male volunteers (age range: 29 to 53 years, mean: 37 years) underwent DW-MRI of the upper abdomen on 1.5T and 3.0T magnetic resonance scanners. Regions of interest in the left and right liver lobe, pancreas, spleen, renal cortex, and renal medulla were delineated independently by two readers. DNNs were trained for IVIM model fitting using these data; results were compared to least-squares and Bayesian approaches to IVIM fitting. Intraclass Correlation Coefficients (ICC) were used to assess consistency of measurements between readers. Intersubject variability was evaluated using Coefficients of Variation (CV). The fitting error was calculated based on simulated data and the average fitting time of each method was recorded. Results: DNNs were trained successfully for IVIM parameter estimation. This approach was associated with high consistency between the two readers (ICCs between 50 and 97%), low intersubject variability of estimated parameter values (CVs between 9.2 and 28.4), and the lowest error when compared with least-squares and Bayesian approaches. Further, fitting by DNNs was several orders of magnitude quicker than the other methods. Conclusion: DNNs are recommended for accurate and robust IVIM model fitting to DW-MRI data. Suitable software is available at (1).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge