Anders Sönnerborg
A graph neural network-based model with Out-of-Distribution Robustness for enhancing Antiretroviral Therapy Outcome Prediction for HIV-1
Dec 29, 2023


Abstract:Predicting the outcome of antiretroviral therapies for HIV-1 is a pressing clinical challenge, especially when the treatment regimen includes drugs for which limited effectiveness data is available. This scarcity of data can arise either due to the introduction of a new drug to the market or due to limited use in clinical settings. To tackle this issue, we introduce a novel joint fusion model, which combines features from a Fully Connected (FC) Neural Network and a Graph Neural Network (GNN). The FC network employs tabular data with a feature vector made up of viral mutations identified in the most recent genotypic resistance test, along with the drugs used in therapy. Conversely, the GNN leverages knowledge derived from Stanford drug-resistance mutation tables, which serve as benchmark references for deducing in-vivo treatment efficacy based on the viral genetic sequence, to build informative graphs. We evaluated these models' robustness against Out-of-Distribution drugs in the test set, with a specific focus on the GNN's role in handling such scenarios. Our comprehensive analysis demonstrates that the proposed model consistently outperforms the FC model, especially when considering Out-of-Distribution drugs. These results underscore the advantage of integrating Stanford scores in the model, thereby enhancing its generalizability and robustness, but also extending its utility in real-world applications with limited data availability. This research highlights the potential of our approach to inform antiretroviral therapy outcome prediction and contribute to more informed clinical decisions.
Incorporating temporal dynamics of mutations to enhance the prediction capability of antiretroviral therapy's outcome for HIV-1
Nov 08, 2023Abstract:Motivation: In predicting HIV therapy outcomes, a critical clinical question is whether using historical information can enhance predictive capabilities compared with current or latest available data analysis. This study analyses whether historical knowledge, which includes viral mutations detected in all genotypic tests before therapy, their temporal occurrence, and concomitant viral load measurements, can bring improvements. We introduce a method to weigh mutations, considering the previously enumerated factors and the reference mutation-drug Stanford resistance tables. We compare a model encompassing history (H) with one not using it (NH). Results: The H-model demonstrates superior discriminative ability, with a higher ROC-AUC score (76.34%) than the NH-model (74.98%). Significant Wilcoxon test results confirm that incorporating historical information improves consistently predictive accuracy for treatment outcomes. The better performance of the H-model might be attributed to its consideration of latent HIV reservoirs, probably obtained when leveraging historical information. The findings emphasize the importance of temporal dynamics in mutations, offering insights into HIV infection complexities. However, our result also shows that prediction accuracy remains relatively high even when no historical information is available. Supplementary information: Supplementary material is available.
The Health Gym: Synthetic Health-Related Datasets for the Development of Reinforcement Learning Algorithms
Mar 12, 2022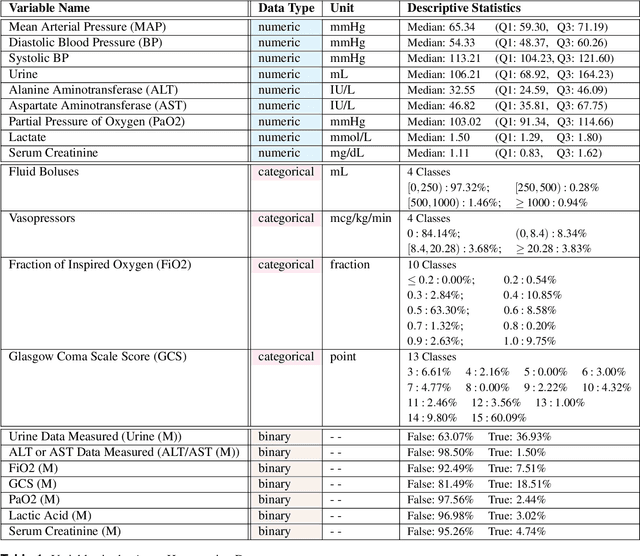
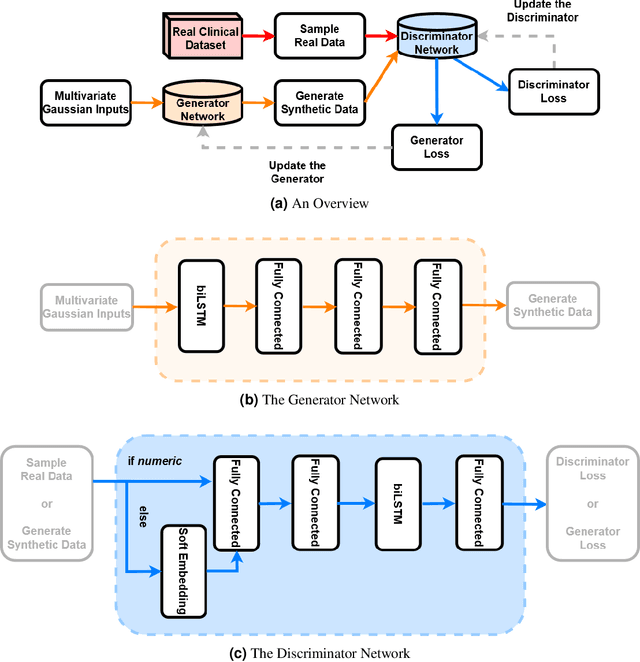
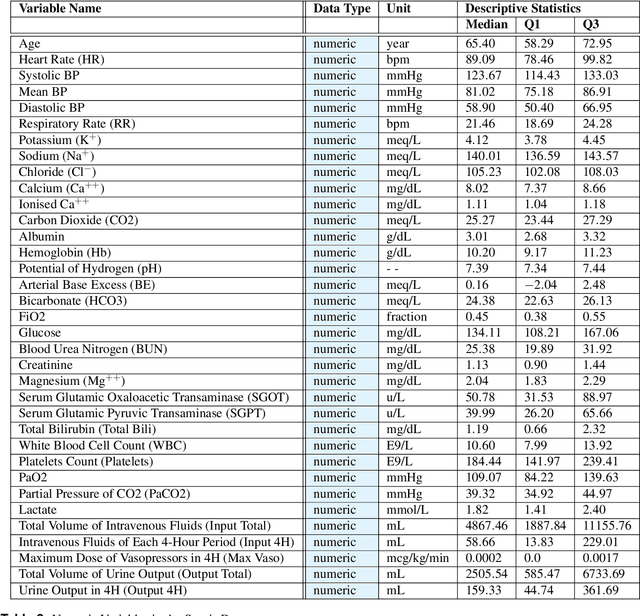
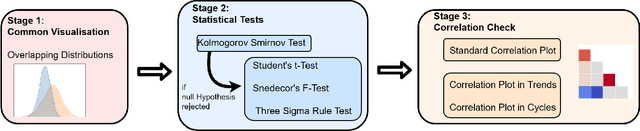
Abstract:In recent years, the machine learning research community has benefited tremendously from the availability of openly accessible benchmark datasets. Clinical data are usually not openly available due to their highly confidential nature. This has hampered the development of reproducible and generalisable machine learning applications in health care. Here we introduce the Health Gym - a growing collection of highly realistic synthetic medical datasets that can be freely accessed to prototype, evaluate, and compare machine learning algorithms, with a specific focus on reinforcement learning. The three synthetic datasets described in this paper present patient cohorts with acute hypotension and sepsis in the intensive care unit, and people with human immunodeficiency virus (HIV) receiving antiretroviral therapy in ambulatory care. The datasets were created using a novel generative adversarial network (GAN). The distributions of variables, and correlations between variables and trends over time in the synthetic datasets mirror those in the real datasets. Furthermore, the risk of sensitive information disclosure associated with the public distribution of the synthetic datasets is estimated to be very low.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge