Valerio Guarrasi
Handling Missing Modalities in Multimodal Survival Prediction for Non-Small Cell Lung Cancer
Jan 15, 2026Abstract:Accurate survival prediction in Non-Small Cell Lung Cancer (NSCLC) requires the integration of heterogeneous clinical, radiological, and histopathological information. While Multimodal Deep Learning (MDL) offers a promises for precision prognosis and survival prediction, its clinical applicability is severely limited by small cohort sizes and the presence of missing modalities, often forcing complete-case filtering or aggressive imputation. In this work, we present a missing-aware multimodal survival framework that integrates Computed Tomography (CT), Whole-Slide Histopathology (WSI) Images, and structured clinical variables for overall survival modeling in unresectable stage II-III NSCLC. By leveraging Foundation Models (FM) for modality-specific feature extraction and a missing-aware encoding strategy, the proposed approach enables intermediate multimodal fusion under naturally incomplete modality profiles. The proposed architecture is resilient to missing modalities by design, allowing the model to utilize all available data without being forced to drop patients during training or inference. Experimental results demonstrate that intermediate fusion consistently outperforms unimodal baselines as well as early and late fusion strategies, with the strongest performance achieved by the fusion of WSI and clinical modalities (73.30 C-index). Further analyses of modality importance reveal an adaptive behavior in which less informative modalities, i.e., CT modality, are automatically down-weighted and contribute less to the final survival prediction.
Sample-Aware Test-Time Adaptation for Medical Image-to-Image Translation
Aug 01, 2025Abstract:Image-to-image translation has emerged as a powerful technique in medical imaging, enabling tasks such as image denoising and cross-modality conversion. However, it suffers from limitations in handling out-of-distribution samples without causing performance degradation. To address this limitation, we propose a novel Test-Time Adaptation (TTA) framework that dynamically adjusts the translation process based on the characteristics of each test sample. Our method introduces a Reconstruction Module to quantify the domain shift and a Dynamic Adaptation Block that selectively modifies the internal features of a pretrained translation model to mitigate the shift without compromising the performance on in-distribution samples that do not require adaptation. We evaluate our approach on two medical image-to-image translation tasks: low-dose CT denoising and T1 to T2 MRI translation, showing consistent improvements over both the baseline translation model without TTA and prior TTA methods. Our analysis highlights the limitations of the state-of-the-art that uniformly apply the adaptation to both out-of-distribution and in-distribution samples, demonstrating that dynamic, sample-specific adjustment offers a promising path to improve model resilience in real-world scenarios. The code is available at: https://github.com/cosbidev/Sample-Aware_TTA.
Leveraging MIMIC Datasets for Better Digital Health: A Review on Open Problems, Progress Highlights, and Future Promises
Jun 15, 2025Abstract:The Medical Information Mart for Intensive Care (MIMIC) datasets have become the Kernel of Digital Health Research by providing freely accessible, deidentified records from tens of thousands of critical care admissions, enabling a broad spectrum of applications in clinical decision support, outcome prediction, and healthcare analytics. Although numerous studies and surveys have explored the predictive power and clinical utility of MIMIC based models, critical challenges in data integration, representation, and interoperability remain underexplored. This paper presents a comprehensive survey that focuses uniquely on open problems. We identify persistent issues such as data granularity, cardinality limitations, heterogeneous coding schemes, and ethical constraints that hinder the generalizability and real-time implementation of machine learning models. We highlight key progress in dimensionality reduction, temporal modelling, causal inference, and privacy preserving analytics, while also outlining promising directions including hybrid modelling, federated learning, and standardized preprocessing pipelines. By critically examining these structural limitations and their implications, this survey offers actionable insights to guide the next generation of MIMIC powered digital health innovations.
Timing Is Everything: Finding the Optimal Fusion Points in Multimodal Medical Imaging
May 05, 2025Abstract:Multimodal deep learning harnesses diverse imaging modalities, such as MRI sequences, to enhance diagnostic accuracy in medical imaging. A key challenge is determining the optimal timing for integrating these modalities-specifically, identifying the network layers where fusion modules should be inserted. Current approaches often rely on manual tuning or exhaustive search, which are computationally expensive without any guarantee of converging to optimal results. We propose a sequential forward search algorithm that incrementally activates and evaluates candidate fusion modules at different layers of a multimodal network. At each step, the algorithm retrains from previously learned weights and compares validation loss to identify the best-performing configuration. This process systematically reduces the search space, enabling efficient identification of the optimal fusion timing without exhaustively testing all possible module placements. The approach is validated on two multimodal MRI datasets, each addressing different classification tasks. Our algorithm consistently identified configurations that outperformed unimodal baselines, late fusion, and a brute-force ensemble of all potential fusion placements. These architectures demonstrated superior accuracy, F-score, and specificity while maintaining competitive or improved AUC values. Furthermore, the sequential nature of the search significantly reduced computational overhead, making the optimization process more practical. By systematically determining the optimal timing to fuse imaging modalities, our method advances multimodal deep learning for medical imaging. It provides an efficient and robust framework for fusion optimization, paving the way for improved clinical decision-making and more adaptable, scalable architectures in medical AI applications.
Lesion-Aware Generative Artificial Intelligence for Virtual Contrast-Enhanced Mammography in Breast Cancer
May 05, 2025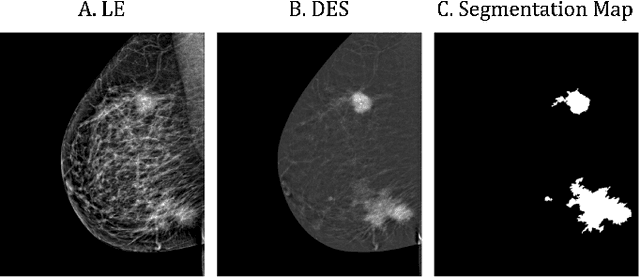
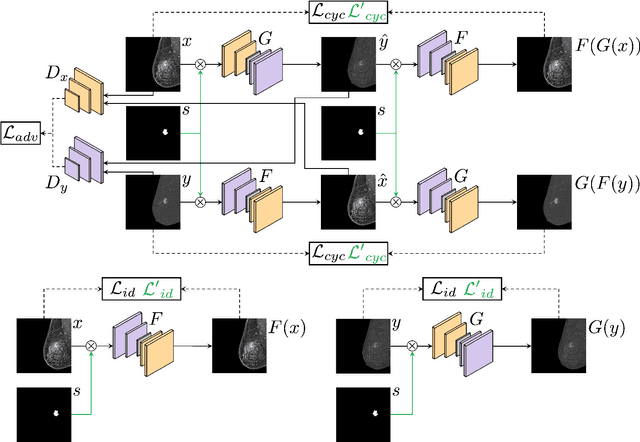
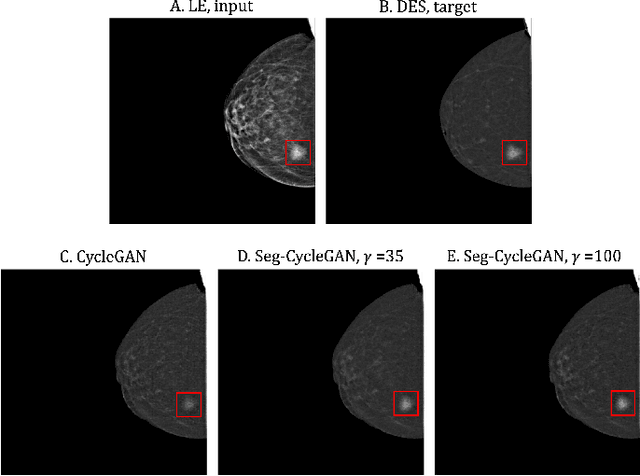
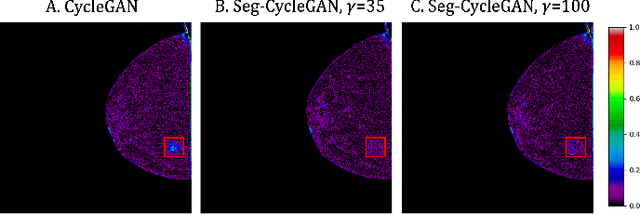
Abstract:Contrast-Enhanced Spectral Mammography (CESM) is a dual-energy mammographic technique that improves lesion visibility through the administration of an iodinated contrast agent. It acquires both a low-energy image, comparable to standard mammography, and a high-energy image, which are then combined to produce a dual-energy subtracted image highlighting lesion contrast enhancement. While CESM offers superior diagnostic accuracy compared to standard mammography, its use entails higher radiation exposure and potential side effects associated with the contrast medium. To address these limitations, we propose Seg-CycleGAN, a generative deep learning framework for Virtual Contrast Enhancement in CESM. The model synthesizes high-fidelity dual-energy subtracted images from low-energy images, leveraging lesion segmentation maps to guide the generative process and improve lesion reconstruction. Building upon the standard CycleGAN architecture, Seg-CycleGAN introduces localized loss terms focused on lesion areas, enhancing the synthesis of diagnostically relevant regions. Experiments on the CESM@UCBM dataset demonstrate that Seg-CycleGAN outperforms the baseline in terms of PSNR and SSIM, while maintaining competitive MSE and VIF. Qualitative evaluations further confirm improved lesion fidelity in the generated images. These results suggest that segmentation-aware generative models offer a viable pathway toward contrast-free CESM alternatives.
Multimodal Doctor-in-the-Loop: A Clinically-Guided Explainable Framework for Predicting Pathological Response in Non-Small Cell Lung Cancer
May 02, 2025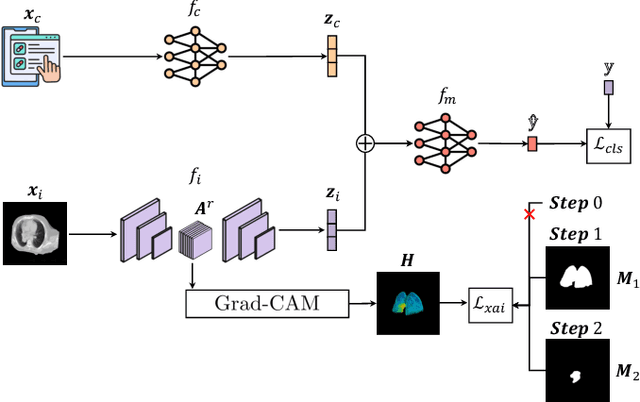
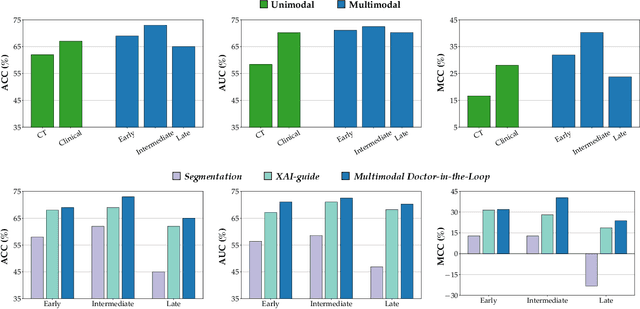
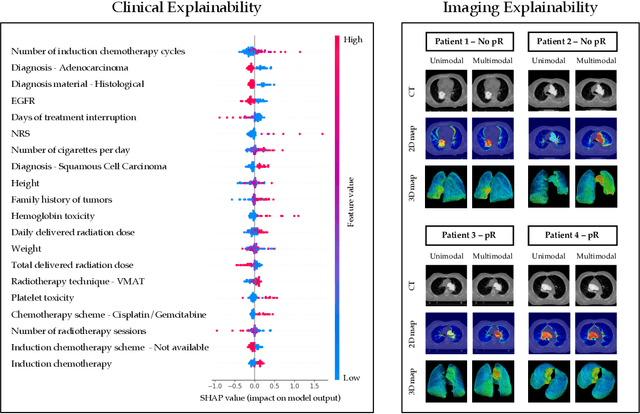
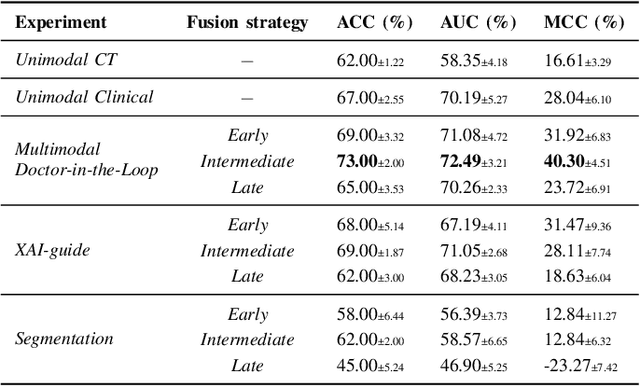
Abstract:This study proposes a novel approach combining Multimodal Deep Learning with intrinsic eXplainable Artificial Intelligence techniques to predict pathological response in non-small cell lung cancer patients undergoing neoadjuvant therapy. Due to the limitations of existing radiomics and unimodal deep learning approaches, we introduce an intermediate fusion strategy that integrates imaging and clinical data, enabling efficient interaction between data modalities. The proposed Multimodal Doctor-in-the-Loop method further enhances clinical relevance by embedding clinicians' domain knowledge directly into the training process, guiding the model's focus gradually from broader lung regions to specific lesions. Results demonstrate improved predictive accuracy and explainability, providing insights into optimal data integration strategies for clinical applications.
Can Foundation Models Really Segment Tumors? A Benchmarking Odyssey in Lung CT Imaging
May 02, 2025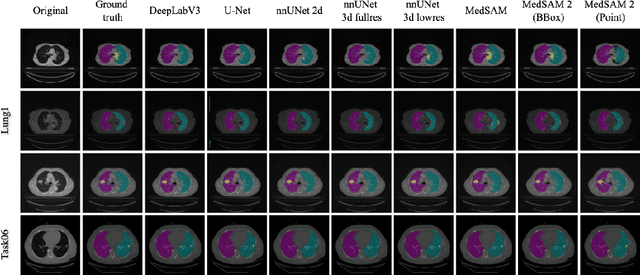
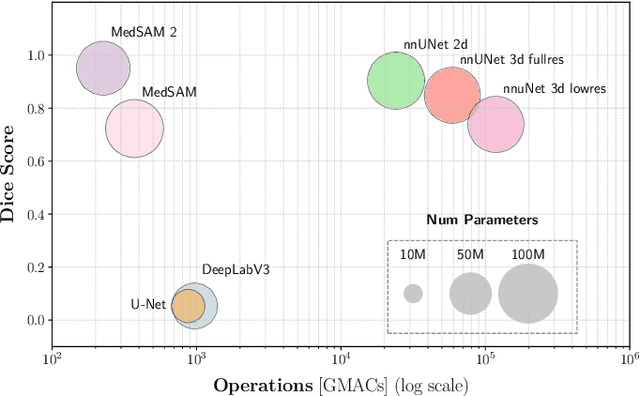
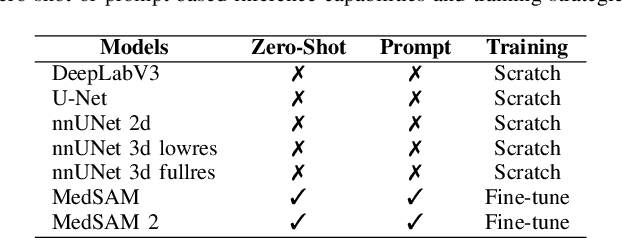
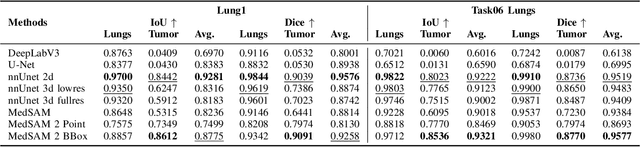
Abstract:Accurate lung tumor segmentation is crucial for improving diagnosis, treatment planning, and patient outcomes in oncology. However, the complexity of tumor morphology, size, and location poses significant challenges for automated segmentation. This study presents a comprehensive benchmarking analysis of deep learning-based segmentation models, comparing traditional architectures such as U-Net and DeepLabV3, self-configuring models like nnUNet, and foundation models like MedSAM, and MedSAM~2. Evaluating performance across two lung tumor segmentation datasets, we assess segmentation accuracy and computational efficiency under various learning paradigms, including few-shot learning and fine-tuning. The results reveal that while traditional models struggle with tumor delineation, foundation models, particularly MedSAM~2, outperform them in both accuracy and computational efficiency. These findings underscore the potential of foundation models for lung tumor segmentation, highlighting their applicability in improving clinical workflows and patient outcomes.
Any-to-Any Vision-Language Model for Multimodal X-ray Imaging and Radiological Report Generation
May 02, 2025Abstract:Generative models have revolutionized Artificial Intelligence (AI), particularly in multimodal applications. However, adapting these models to the medical domain poses unique challenges due to the complexity of medical data and the stringent need for clinical accuracy. In this work, we introduce a framework specifically designed for multimodal medical data generation. By enabling the generation of multi-view chest X-rays and their associated clinical report, it bridges the gap between general-purpose vision-language models and the specialized requirements of healthcare. Leveraging the MIMIC-CXR dataset, the proposed framework shows superior performance in generating high-fidelity images and semantically coherent reports. Our quantitative evaluation reveals significant results in terms of FID and BLEU scores, showcasing the quality of the generated data. Notably, our framework achieves comparable or even superior performance compared to real data on downstream disease classification tasks, underlining its potential as a tool for medical research and diagnostics. This study highlights the importance of domain-specific adaptations in enhancing the relevance and utility of generative models for clinical applications, paving the way for future advancements in synthetic multimodal medical data generation.
Evaluating Vision Language Model Adaptations for Radiology Report Generation in Low-Resource Languages
May 02, 2025Abstract:The integration of artificial intelligence in healthcare has opened new horizons for improving medical diagnostics and patient care. However, challenges persist in developing systems capable of generating accurate and contextually relevant radiology reports, particularly in low-resource languages. In this study, we present a comprehensive benchmark to evaluate the performance of instruction-tuned Vision-Language Models (VLMs) in the specialized task of radiology report generation across three low-resource languages: Italian, German, and Spanish. Employing the LLaVA architectural framework, we conducted a systematic evaluation of pre-trained models utilizing general datasets, domain-specific datasets, and low-resource language-specific datasets. In light of the unavailability of models that possess prior knowledge of both the medical domain and low-resource languages, we analyzed various adaptations to determine the most effective approach for these contexts. The results revealed that language-specific models substantially outperformed both general and domain-specific models in generating radiology reports, emphasizing the critical role of linguistic adaptation. Additionally, models fine-tuned with medical terminology exhibited enhanced performance across all languages compared to models with generic knowledge, highlighting the importance of domain-specific training. We also explored the influence of the temperature parameter on the coherence of report generation, providing insights for optimal model settings. Our findings highlight the importance of tailored language and domain-specific training for improving the quality and accuracy of radiological reports in multilingual settings. This research not only advances our understanding of VLMs adaptability in healthcare but also points to significant avenues for future investigations into model tuning and language-specific adaptations.
Beyond the Generative Learning Trilemma: Generative Model Assessment in Data Scarcity Domains
Apr 14, 2025
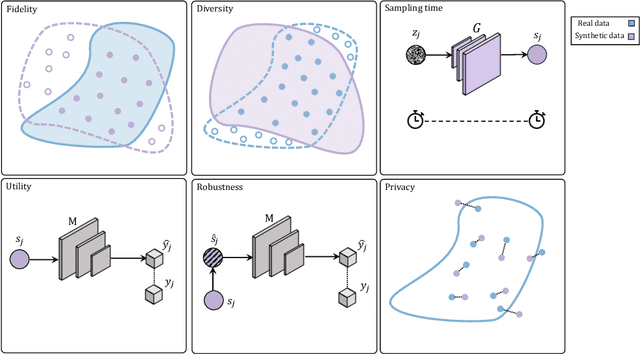
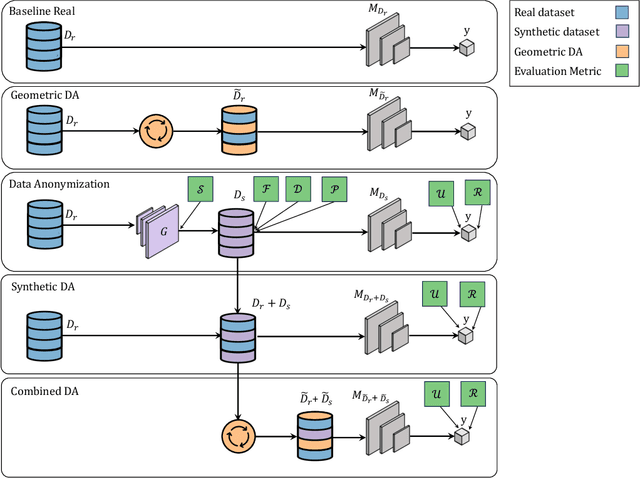
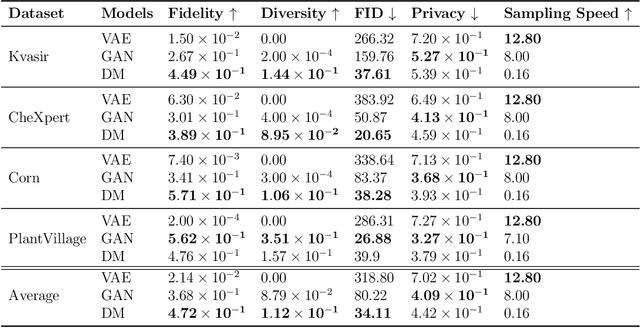
Abstract:Data scarcity remains a critical bottleneck impeding technological advancements across various domains, including but not limited to medicine and precision agriculture. To address this challenge, we explore the potential of Deep Generative Models (DGMs) in producing synthetic data that satisfies the Generative Learning Trilemma: fidelity, diversity, and sampling efficiency. However, recognizing that these criteria alone are insufficient for practical applications, we extend the trilemma to include utility, robustness, and privacy, factors crucial for ensuring the applicability of DGMs in real-world scenarios. Evaluating these metrics becomes particularly challenging in data-scarce environments, as DGMs traditionally rely on large datasets to perform optimally. This limitation is especially pronounced in domains like medicine and precision agriculture, where ensuring acceptable model performance under data constraints is vital. To address these challenges, we assess the Generative Learning Trilemma in data-scarcity settings using state-of-the-art evaluation metrics, comparing three prominent DGMs: Variational Autoencoders (VAEs), Generative Adversarial Networks (GANs), and Diffusion Models (DMs). Furthermore, we propose a comprehensive framework to assess utility, robustness, and privacy in synthetic data generated by DGMs. Our findings demonstrate varying strengths among DGMs, with each model exhibiting unique advantages based on the application context. This study broadens the scope of the Generative Learning Trilemma, aligning it with real-world demands and providing actionable guidance for selecting DGMs tailored to specific applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge