Sara Ramella
Handling Missing Modalities in Multimodal Survival Prediction for Non-Small Cell Lung Cancer
Jan 15, 2026Abstract:Accurate survival prediction in Non-Small Cell Lung Cancer (NSCLC) requires the integration of heterogeneous clinical, radiological, and histopathological information. While Multimodal Deep Learning (MDL) offers a promises for precision prognosis and survival prediction, its clinical applicability is severely limited by small cohort sizes and the presence of missing modalities, often forcing complete-case filtering or aggressive imputation. In this work, we present a missing-aware multimodal survival framework that integrates Computed Tomography (CT), Whole-Slide Histopathology (WSI) Images, and structured clinical variables for overall survival modeling in unresectable stage II-III NSCLC. By leveraging Foundation Models (FM) for modality-specific feature extraction and a missing-aware encoding strategy, the proposed approach enables intermediate multimodal fusion under naturally incomplete modality profiles. The proposed architecture is resilient to missing modalities by design, allowing the model to utilize all available data without being forced to drop patients during training or inference. Experimental results demonstrate that intermediate fusion consistently outperforms unimodal baselines as well as early and late fusion strategies, with the strongest performance achieved by the fusion of WSI and clinical modalities (73.30 C-index). Further analyses of modality importance reveal an adaptive behavior in which less informative modalities, i.e., CT modality, are automatically down-weighted and contribute less to the final survival prediction.
Multimodal Doctor-in-the-Loop: A Clinically-Guided Explainable Framework for Predicting Pathological Response in Non-Small Cell Lung Cancer
May 02, 2025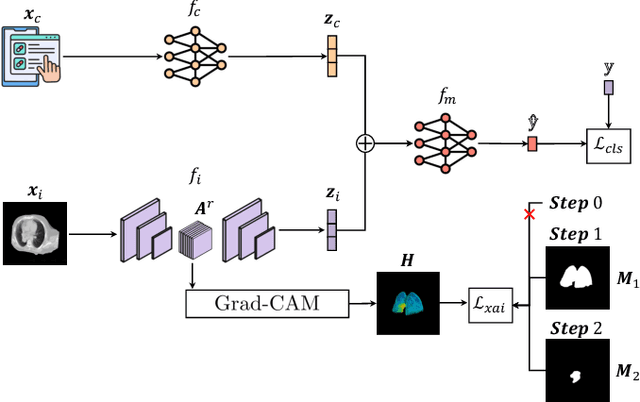
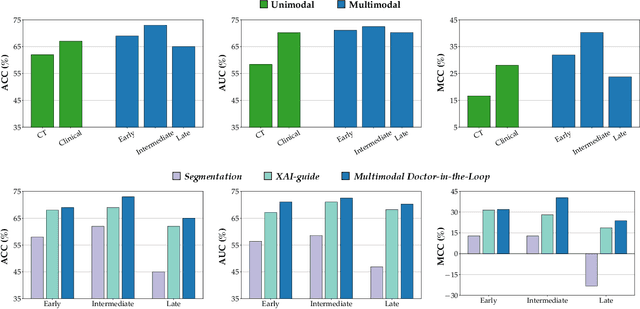
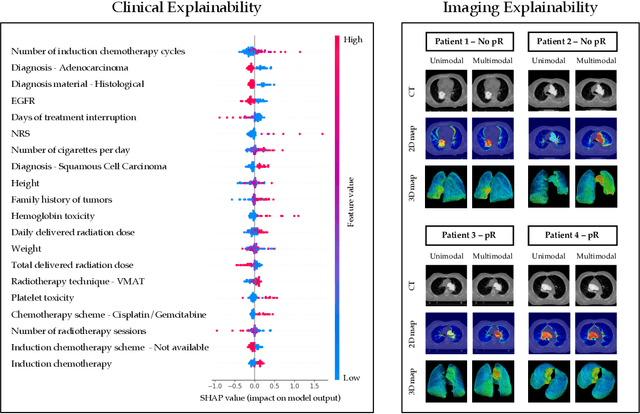
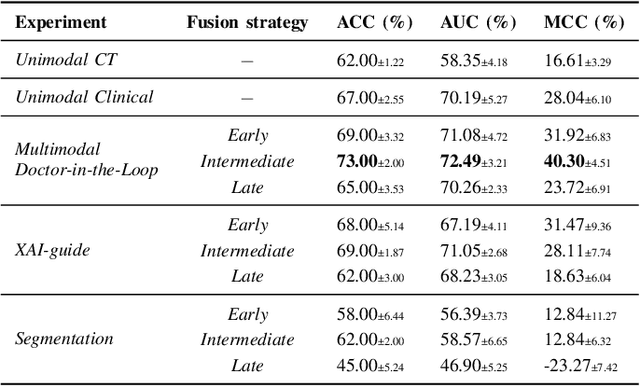
Abstract:This study proposes a novel approach combining Multimodal Deep Learning with intrinsic eXplainable Artificial Intelligence techniques to predict pathological response in non-small cell lung cancer patients undergoing neoadjuvant therapy. Due to the limitations of existing radiomics and unimodal deep learning approaches, we introduce an intermediate fusion strategy that integrates imaging and clinical data, enabling efficient interaction between data modalities. The proposed Multimodal Doctor-in-the-Loop method further enhances clinical relevance by embedding clinicians' domain knowledge directly into the training process, guiding the model's focus gradually from broader lung regions to specific lesions. Results demonstrate improved predictive accuracy and explainability, providing insights into optimal data integration strategies for clinical applications.
Doctor-in-the-Loop: An Explainable, Multi-View Deep Learning Framework for Predicting Pathological Response in Non-Small Cell Lung Cancer
Feb 21, 2025Abstract:Non-small cell lung cancer (NSCLC) remains a major global health challenge, with high post-surgical recurrence rates underscoring the need for accurate pathological response predictions to guide personalized treatments. Although artificial intelligence models show promise in this domain, their clinical adoption is limited by the lack of medically grounded guidance during training, often resulting in non-explainable intrinsic predictions. To address this, we propose Doctor-in-the-Loop, a novel framework that integrates expert-driven domain knowledge with explainable artificial intelligence techniques, directing the model toward clinically relevant anatomical regions and improving both interpretability and trustworthiness. Our approach employs a gradual multi-view strategy, progressively refining the model's focus from broad contextual features to finer, lesion-specific details. By incorporating domain insights at every stage, we enhance predictive accuracy while ensuring that the model's decision-making process aligns more closely with clinical reasoning. Evaluated on a dataset of NSCLC patients, Doctor-in-the-Loop delivers promising predictive performance and provides transparent, justifiable outputs, representing a significant step toward clinically explainable artificial intelligence in oncology.
A Deep Learning Approach for Overall Survival Prediction in Lung Cancer with Missing Values
Jul 28, 2023Abstract:One of the most challenging fields where Artificial Intelligence (AI) can be applied is lung cancer research, specifically non-small cell lung cancer (NSCLC). In particular, overall survival (OS), the time between diagnosis and death, is a vital indicator of patient status, enabling tailored treatment and improved OS rates. In this analysis, there are two challenges to take into account. First, few studies effectively exploit the information available from each patient, leveraging both uncensored (i.e., dead) and censored (i.e., survivors) patients, considering also the events' time. Second, the handling of incomplete data is a common issue in the medical field. This problem is typically tackled through the use of imputation methods. Our objective is to present an AI model able to overcome these limits, effectively learning from both censored and uncensored patients and their available features, for the prediction of OS for NSCLC patients. We present a novel approach to survival analysis with missing values in the context of NSCLC, which exploits the strengths of the transformer architecture to account only for available features without requiring any imputation strategy. By making use of ad-hoc losses for OS, it is able to account for both censored and uncensored patients, as well as changes in risks over time. We compared our method with state-of-the-art models for survival analysis coupled with different imputation strategies. We evaluated the results obtained over a period of 6 years using different time granularities obtaining a Ct-index, a time-dependent variant of the C-index, of 71.97, 77.58 and 80.72 for time units of 1 month, 1 year and 2 years, respectively, outperforming all state-of-the-art methods regardless of the imputation method used.
RadioPathomics: Multimodal Learning in Non-Small Cell Lung Cancer for Adaptive Radiotherapy
Apr 26, 2022



Abstract:The current cancer treatment practice collects multimodal data, such as radiology images, histopathology slides, genomics and clinical data. The importance of these data sources taken individually has fostered the recent raise of radiomics and pathomics, i.e. the extraction of quantitative features from radiology and histopathology images routinely collected to predict clinical outcomes or to guide clinical decisions using artificial intelligence algorithms. Nevertheless, how to combine them into a single multimodal framework is still an open issue. In this work we therefore develop a multimodal late fusion approach that combines hand-crafted features computed from radiomics, pathomics and clinical data to predict radiation therapy treatment outcomes for non-small-cell lung cancer patients. Within this context, we investigate eight different late fusion rules (i.e. product, maximum, minimum, mean, decision template, Dempster-Shafer, majority voting, and confidence rule) and two patient-wise aggregation rules leveraging the richness of information given by computer tomography images and whole-slide scans. The experiments in leave-one-patient-out cross-validation on an in-house cohort of 33 patients show that the proposed multimodal paradigm with an AUC equal to $90.9\%$ outperforms each unimodal approach, suggesting that data integration can advance precision medicine. As a further contribution, we also compare the hand-crafted representations with features automatically computed by deep networks, and the late fusion paradigm with early fusion, another popular multimodal approach. In both cases, the experiments show that the proposed multimodal approach provides the best results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge