Nicholas I-Hsien Kuo
Limits of Generative Pre-Training in Structured EMR Trajectories with Irregular Sampling
Oct 27, 2025Abstract:Foundation models refer to architectures trained on vast datasets using autoregressive pre-training from natural language processing to capture intricate patterns and motifs. They were originally developed to transfer such learned knowledge to downstream predictive tasks. Recently, however, some studies repurpose these learned representations for phenotype discovery without rigorous validation, risking superficially realistic but clinically incoherent embeddings. To test this mismatch, we trained two autoregressive models -- a sequence-to-sequence LSTM and a reduced Transformer -- on longitudinal ART for HIV and Acute Hypotension datasets. Controlled irregularity was added during training via random inter-visit gaps, while test sequences stayed complete. Patient-trajectory synthesis evaluated distributional and correlational fidelity. Both reproduced feature distributions but failed to preserve cross-feature structure -- showing that generative pre-training yields local realism but limited clinical coherence. These results highlight the need for domain-specific evaluation and support trajectory synthesis as a practical probe before fine-tuning or deployment.
Attention-Based Synthetic Data Generation for Calibration-Enhanced Survival Analysis: A Case Study for Chronic Kidney Disease Using Electronic Health Records
Mar 08, 2025Abstract:Access to real-world healthcare data is limited by stringent privacy regulations and data imbalances, hindering advancements in research and clinical applications. Synthetic data presents a promising solution, yet existing methods often fail to ensure the realism, utility, and calibration essential for robust survival analysis. Here, we introduce Masked Clinical Modelling (MCM), an attention-based framework capable of generating high-fidelity synthetic datasets that preserve critical clinical insights, such as hazard ratios, while enhancing survival model calibration. Unlike traditional statistical methods like SMOTE and machine learning models such as VAEs, MCM supports both standalone dataset synthesis for reproducibility and conditional simulation for targeted augmentation, addressing diverse research needs. Validated on a chronic kidney disease electronic health records dataset, MCM reduced the general calibration loss over the entire dataset by 15%; and MCM reduced a mean calibration loss by 9% across 10 clinically stratified subgroups, outperforming 15 alternative methods. By bridging data accessibility with translational utility, MCM advances the precision of healthcare models, promoting more efficient use of scarce healthcare resources.
Masked Clinical Modelling: A Framework for Synthetic and Augmented Survival Data Generation
Oct 23, 2024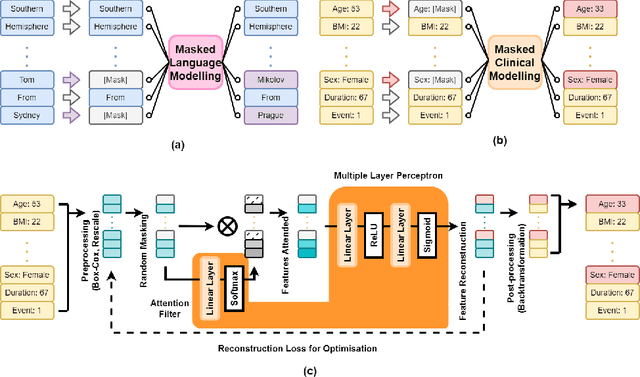
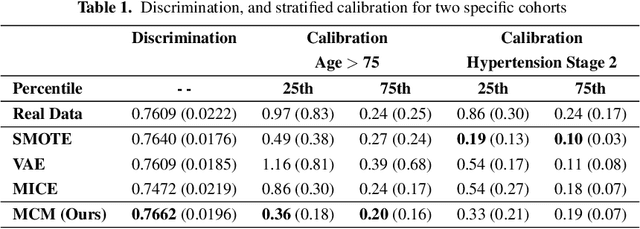
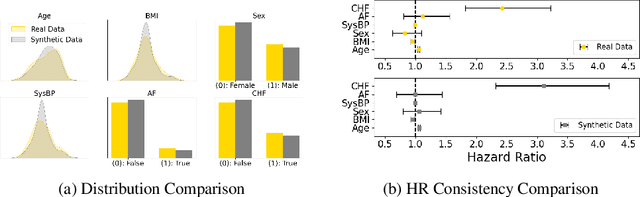
Abstract:Access to real clinical data is often restricted due to privacy obligations, creating significant barriers for healthcare research. Synthetic datasets provide a promising solution, enabling secure data sharing and model development. However, most existing approaches focus on data realism rather than utility -- ensuring that models trained on synthetic data yield clinically meaningful insights comparable to those trained on real data. In this paper, we present Masked Clinical Modelling (MCM), a framework inspired by masked language modelling, designed for both data synthesis and conditional data augmentation. We evaluate this prototype on the WHAS500 dataset using Cox Proportional Hazards models, focusing on the preservation of hazard ratios as key clinical metrics. Our results show that data generated using the MCM framework improves both discrimination and calibration in survival analysis, outperforming existing methods. MCM demonstrates strong potential to support survival data analysis and broader healthcare applications.
CK4Gen: A Knowledge Distillation Framework for Generating High-Utility Synthetic Survival Datasets in Healthcare
Oct 22, 2024Abstract:Access to real clinical data is heavily restricted by privacy regulations, hindering both healthcare research and education. These constraints slow progress in developing new treatments and data-driven healthcare solutions, while also limiting students' access to real-world datasets, leaving them without essential practical skills. High-utility synthetic datasets are therefore critical for advancing research and providing meaningful training material. However, current generative models -- such as Variational Autoencoders (VAEs) and Generative Adversarial Networks (GANs) -- produce surface-level realism at the expense of healthcare utility, blending distinct patient profiles and producing synthetic data of limited practical relevance. To overcome these limitations, we introduce CK4Gen (Cox Knowledge for Generation), a novel framework that leverages knowledge distillation from Cox Proportional Hazards (CoxPH) models to create synthetic survival datasets that preserve key clinical characteristics, including hazard ratios and survival curves. CK4Gen avoids the interpolation issues seen in VAEs and GANs by maintaining distinct patient risk profiles, ensuring realistic and reliable outputs for research and educational use. Validated across four benchmark datasets -- GBSG2, ACTG320, WHAS500, and FLChain -- CK4Gen outperforms competing techniques by better aligning real and synthetic data, enhancing survival model performance in both discrimination and calibration via data augmentation. As CK4Gen is scalable across clinical conditions, and with code to be made publicly available, future researchers can apply it to their own datasets to generate synthetic versions suitable for open sharing.
Synthetic Health-related Longitudinal Data with Mixed-type Variables Generated using Diffusion Models
Mar 22, 2023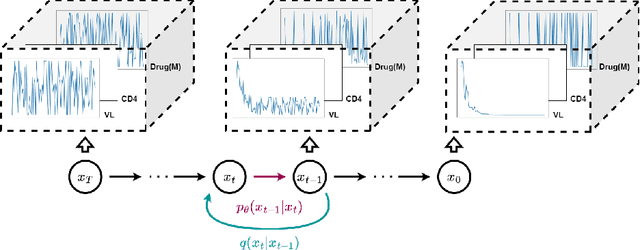

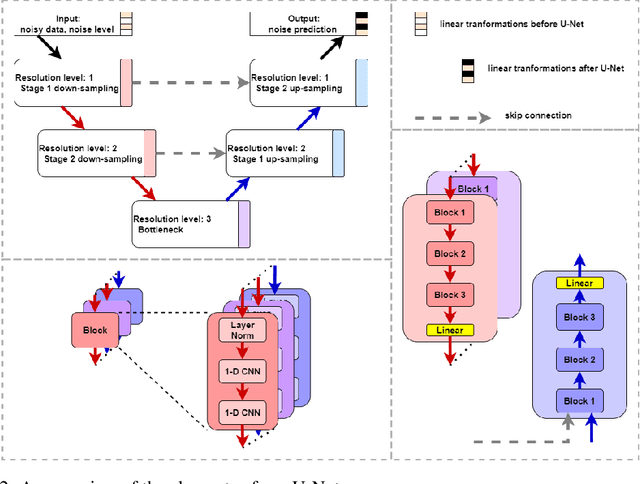
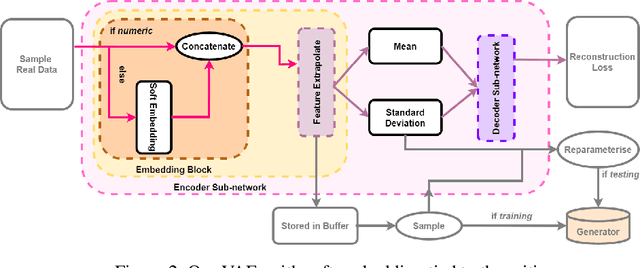
Abstract:This paper presents a novel approach to simulating electronic health records (EHRs) using diffusion probabilistic models (DPMs). Specifically, we demonstrate the effectiveness of DPMs in synthesising longitudinal EHRs that capture mixed-type variables, including numeric, binary, and categorical variables. To our knowledge, this represents the first use of DPMs for this purpose. We compared our DPM-simulated datasets to previous state-of-the-art results based on generative adversarial networks (GANs) for two clinical applications: acute hypotension and human immunodeficiency virus (ART for HIV). Given the lack of similar previous studies in DPMs, a core component of our work involves exploring the advantages and caveats of employing DPMs across a wide range of aspects. In addition to assessing the realism of the synthetic datasets, we also trained reinforcement learning (RL) agents on the synthetic data to evaluate their utility for supporting the development of downstream machine learning models. Finally, we estimated that our DPM-simulated datasets are secure and posed a low patient exposure risk for public access.
Generating Synthetic Clinical Data that Capture Class Imbalanced Distributions with Generative Adversarial Networks: Example using Antiretroviral Therapy for HIV
Aug 18, 2022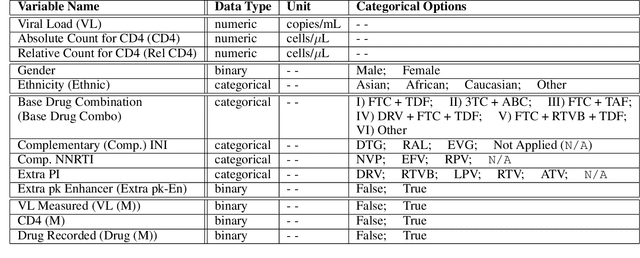
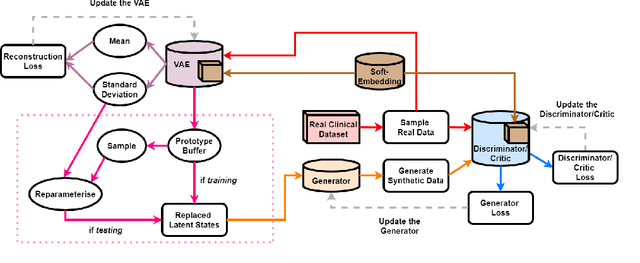
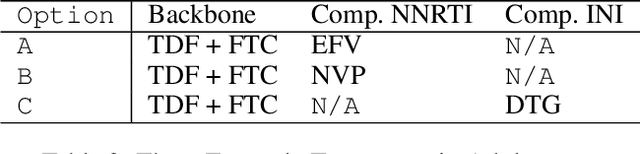
Abstract:Clinical data usually cannot be freely distributed due to their highly confidential nature and this hampers the development of machine learning in the healthcare domain. One way to mitigate this problem is by generating realistic synthetic datasets using generative adversarial networks (GANs). However, GANs are known to suffer from mode collapse and thus creating outputs of low diveristy. In this paper, we extend the classic GAN setup with an external memory to replay features from real samples. Using antiretroviral therapy for human immunodeficiency virus (ART for HIV) as a case study, we show that our extended setup increases convergence and more importantly, it is effective in capturing the severe class imbalanced distributions common to real world clinical data.
The Health Gym: Synthetic Health-Related Datasets for the Development of Reinforcement Learning Algorithms
Mar 12, 2022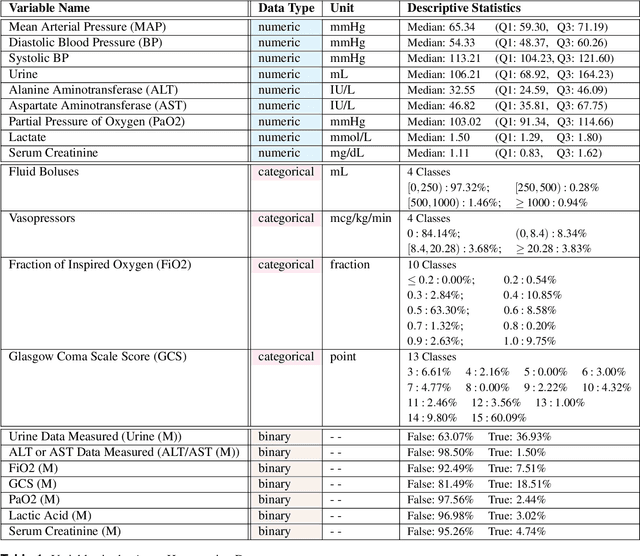
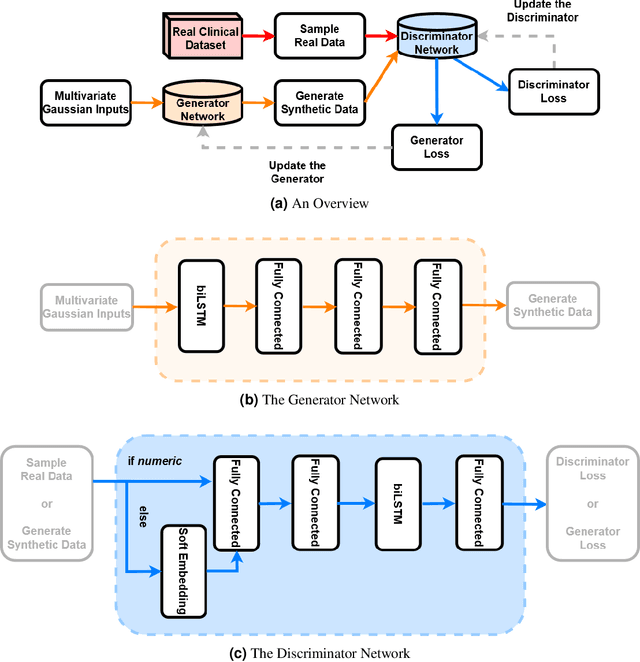
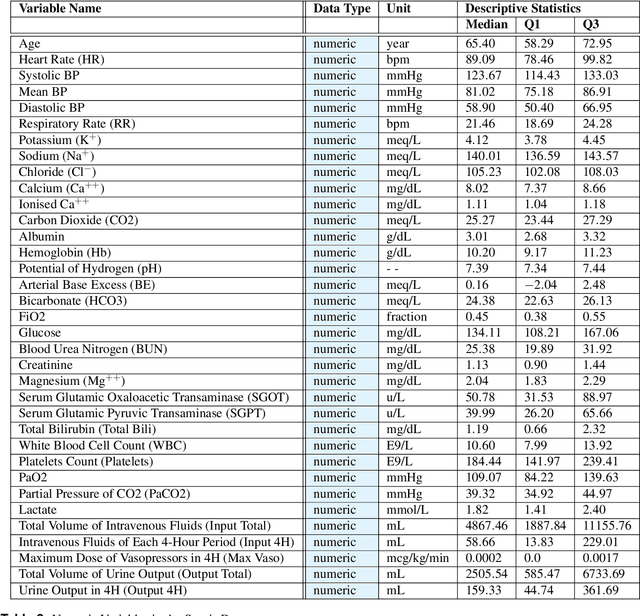
Abstract:In recent years, the machine learning research community has benefited tremendously from the availability of openly accessible benchmark datasets. Clinical data are usually not openly available due to their highly confidential nature. This has hampered the development of reproducible and generalisable machine learning applications in health care. Here we introduce the Health Gym - a growing collection of highly realistic synthetic medical datasets that can be freely accessed to prototype, evaluate, and compare machine learning algorithms, with a specific focus on reinforcement learning. The three synthetic datasets described in this paper present patient cohorts with acute hypotension and sepsis in the intensive care unit, and people with human immunodeficiency virus (HIV) receiving antiretroviral therapy in ambulatory care. The datasets were created using a novel generative adversarial network (GAN). The distributions of variables, and correlations between variables and trends over time in the synthetic datasets mirror those in the real datasets. Furthermore, the risk of sensitive information disclosure associated with the public distribution of the synthetic datasets is estimated to be very low.
Synthetic Acute Hypotension and Sepsis Datasets Based on MIMIC-III and Published as Part of the Health Gym Project
Dec 07, 2021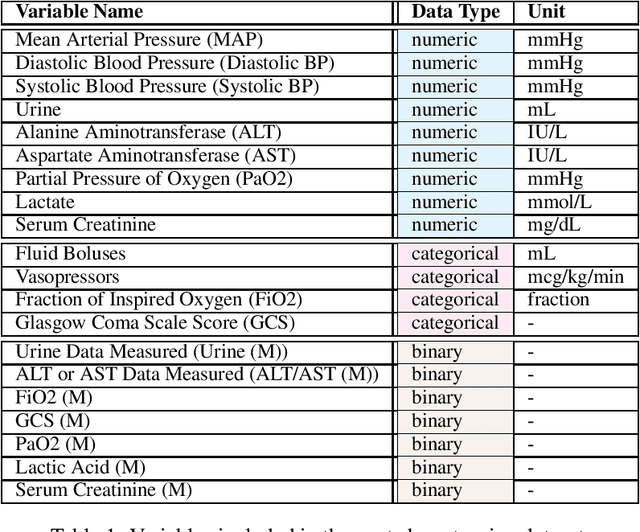

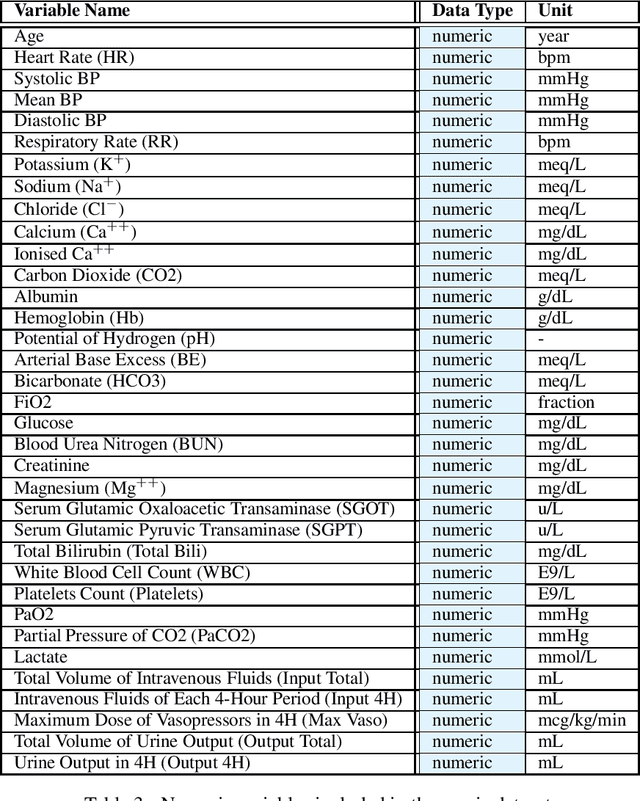
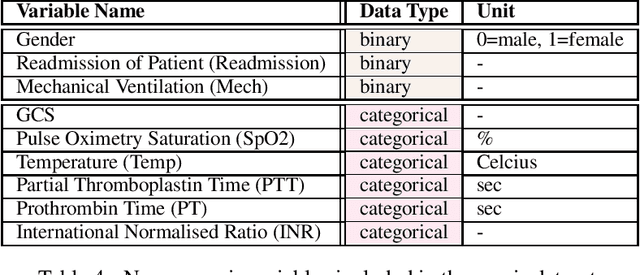
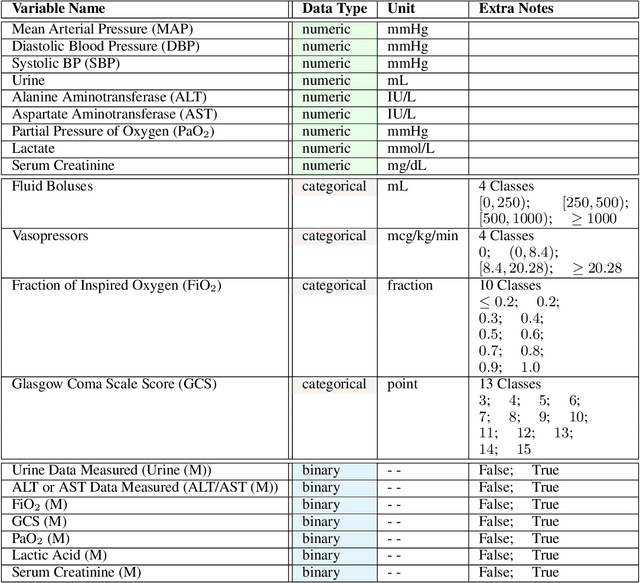
Abstract:These two synthetic datasets comprise vital signs, laboratory test results, administered fluid boluses and vasopressors for 3,910 patients with acute hypotension and for 2,164 patients with sepsis in the Intensive Care Unit (ICU). The patient cohorts were built using previously published inclusion and exclusion criteria and the data were created using Generative Adversarial Networks (GANs) and the MIMIC-III Clinical Database. The risk of identity disclosure associated with the release of these data was estimated to be very low (0.045%). The datasets were generated and published as part of the Health Gym, a project aiming to publicly distribute synthetic longitudinal health data for developing machine learning algorithms (with a particular focus on offline reinforcement learning) and for educational purposes.
Learning to Continually Learn Rapidly from Few and Noisy Data
Mar 06, 2021



Abstract:Neural networks suffer from catastrophic forgetting and are unable to sequentially learn new tasks without guaranteed stationarity in data distribution. Continual learning could be achieved via replay -- by concurrently training externally stored old data while learning a new task. However, replay becomes less effective when each past task is allocated with less memory. To overcome this difficulty, we supplemented replay mechanics with meta-learning for rapid knowledge acquisition. By employing a meta-learner, which \textit{learns a learning rate per parameter per past task}, we found that base learners produced strong results when less memory was available. Additionally, our approach inherited several meta-learning advantages for continual learning: it demonstrated strong robustness to continually learn under the presence of noises and yielded base learners to higher accuracy in less updates.
MTL2L: A Context Aware Neural Optimiser
Jul 18, 2020
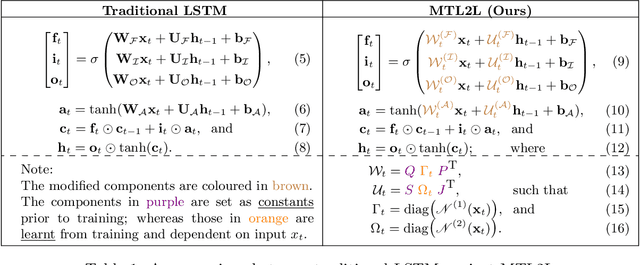

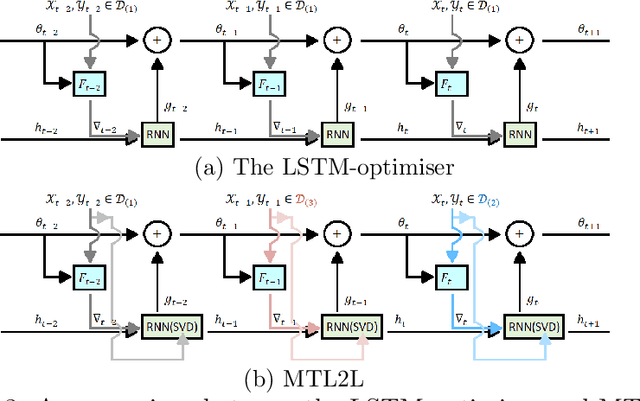
Abstract:Learning to learn (L2L) trains a meta-learner to assist the learning of a task-specific base learner. Previously, it was shown that a meta-learner could learn the direct rules to update learner parameters; and that the learnt neural optimiser updated learners more rapidly than handcrafted gradient-descent methods. However, we demonstrate that previous neural optimisers were limited to update learners on one designated dataset. In order to address input-domain heterogeneity, we introduce Multi-Task Learning to Learn (MTL2L), a context aware neural optimiser which self-modifies its optimisation rules based on input data. We show that MTL2L is capable of updating learners to classify on data of an unseen input-domain at the meta-testing phase.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge