Blanca Gallego
Limits of Generative Pre-Training in Structured EMR Trajectories with Irregular Sampling
Oct 27, 2025Abstract:Foundation models refer to architectures trained on vast datasets using autoregressive pre-training from natural language processing to capture intricate patterns and motifs. They were originally developed to transfer such learned knowledge to downstream predictive tasks. Recently, however, some studies repurpose these learned representations for phenotype discovery without rigorous validation, risking superficially realistic but clinically incoherent embeddings. To test this mismatch, we trained two autoregressive models -- a sequence-to-sequence LSTM and a reduced Transformer -- on longitudinal ART for HIV and Acute Hypotension datasets. Controlled irregularity was added during training via random inter-visit gaps, while test sequences stayed complete. Patient-trajectory synthesis evaluated distributional and correlational fidelity. Both reproduced feature distributions but failed to preserve cross-feature structure -- showing that generative pre-training yields local realism but limited clinical coherence. These results highlight the need for domain-specific evaluation and support trajectory synthesis as a practical probe before fine-tuning or deployment.
Attention-Based Synthetic Data Generation for Calibration-Enhanced Survival Analysis: A Case Study for Chronic Kidney Disease Using Electronic Health Records
Mar 08, 2025Abstract:Access to real-world healthcare data is limited by stringent privacy regulations and data imbalances, hindering advancements in research and clinical applications. Synthetic data presents a promising solution, yet existing methods often fail to ensure the realism, utility, and calibration essential for robust survival analysis. Here, we introduce Masked Clinical Modelling (MCM), an attention-based framework capable of generating high-fidelity synthetic datasets that preserve critical clinical insights, such as hazard ratios, while enhancing survival model calibration. Unlike traditional statistical methods like SMOTE and machine learning models such as VAEs, MCM supports both standalone dataset synthesis for reproducibility and conditional simulation for targeted augmentation, addressing diverse research needs. Validated on a chronic kidney disease electronic health records dataset, MCM reduced the general calibration loss over the entire dataset by 15%; and MCM reduced a mean calibration loss by 9% across 10 clinically stratified subgroups, outperforming 15 alternative methods. By bridging data accessibility with translational utility, MCM advances the precision of healthcare models, promoting more efficient use of scarce healthcare resources.
Masked Clinical Modelling: A Framework for Synthetic and Augmented Survival Data Generation
Oct 23, 2024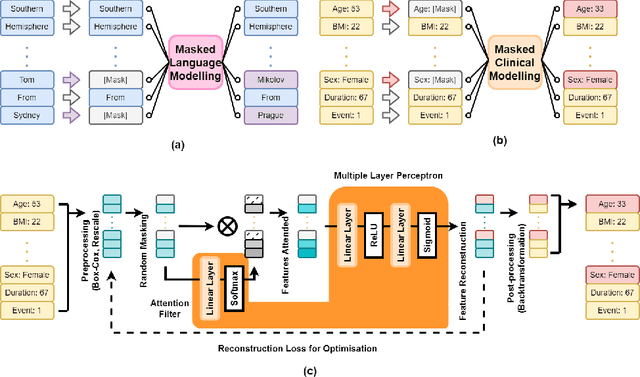
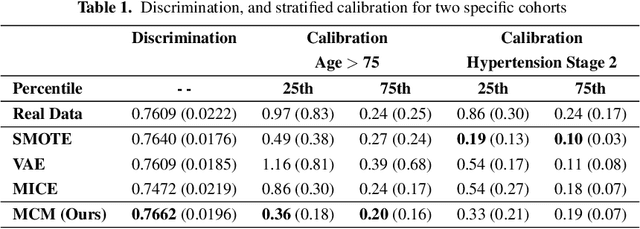
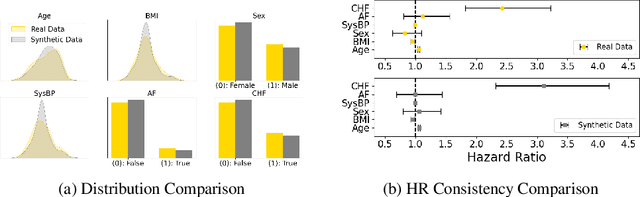
Abstract:Access to real clinical data is often restricted due to privacy obligations, creating significant barriers for healthcare research. Synthetic datasets provide a promising solution, enabling secure data sharing and model development. However, most existing approaches focus on data realism rather than utility -- ensuring that models trained on synthetic data yield clinically meaningful insights comparable to those trained on real data. In this paper, we present Masked Clinical Modelling (MCM), a framework inspired by masked language modelling, designed for both data synthesis and conditional data augmentation. We evaluate this prototype on the WHAS500 dataset using Cox Proportional Hazards models, focusing on the preservation of hazard ratios as key clinical metrics. Our results show that data generated using the MCM framework improves both discrimination and calibration in survival analysis, outperforming existing methods. MCM demonstrates strong potential to support survival data analysis and broader healthcare applications.
CK4Gen: A Knowledge Distillation Framework for Generating High-Utility Synthetic Survival Datasets in Healthcare
Oct 22, 2024Abstract:Access to real clinical data is heavily restricted by privacy regulations, hindering both healthcare research and education. These constraints slow progress in developing new treatments and data-driven healthcare solutions, while also limiting students' access to real-world datasets, leaving them without essential practical skills. High-utility synthetic datasets are therefore critical for advancing research and providing meaningful training material. However, current generative models -- such as Variational Autoencoders (VAEs) and Generative Adversarial Networks (GANs) -- produce surface-level realism at the expense of healthcare utility, blending distinct patient profiles and producing synthetic data of limited practical relevance. To overcome these limitations, we introduce CK4Gen (Cox Knowledge for Generation), a novel framework that leverages knowledge distillation from Cox Proportional Hazards (CoxPH) models to create synthetic survival datasets that preserve key clinical characteristics, including hazard ratios and survival curves. CK4Gen avoids the interpolation issues seen in VAEs and GANs by maintaining distinct patient risk profiles, ensuring realistic and reliable outputs for research and educational use. Validated across four benchmark datasets -- GBSG2, ACTG320, WHAS500, and FLChain -- CK4Gen outperforms competing techniques by better aligning real and synthetic data, enhancing survival model performance in both discrimination and calibration via data augmentation. As CK4Gen is scalable across clinical conditions, and with code to be made publicly available, future researchers can apply it to their own datasets to generate synthetic versions suitable for open sharing.
Predicting adverse outcomes following catheter ablation treatment for atrial fibrillation
Nov 22, 2022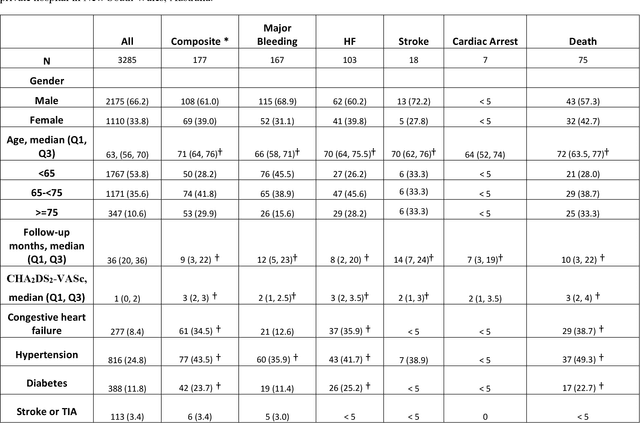
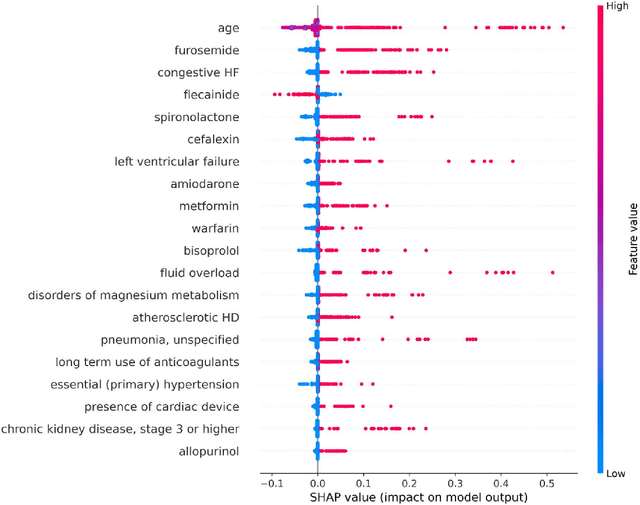
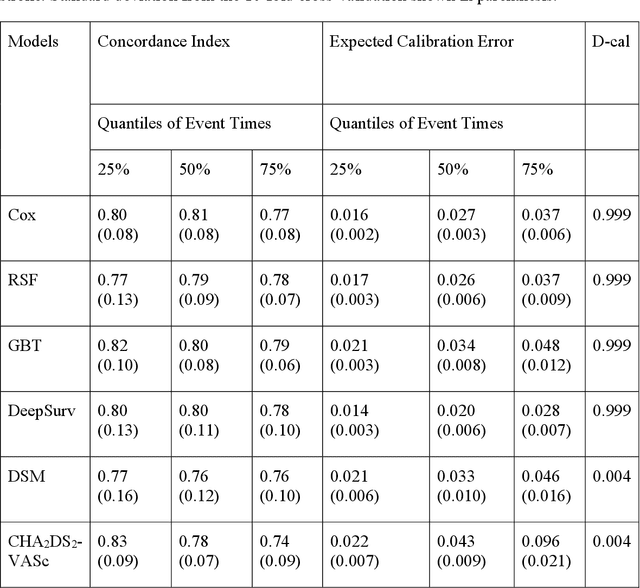
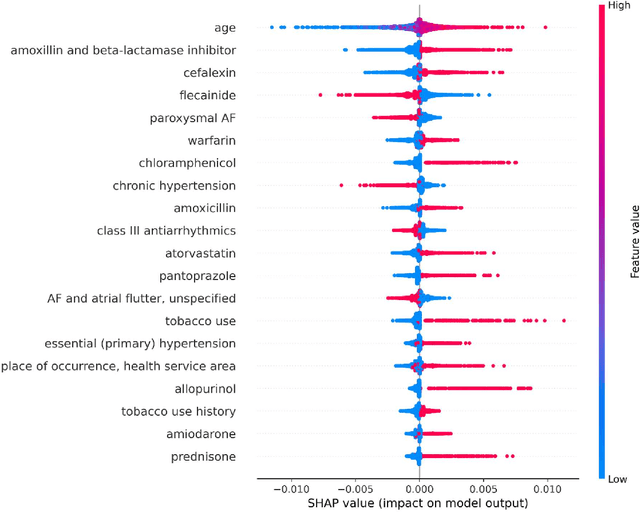
Abstract:Objective: To develop prognostic survival models for predicting adverse outcomes after catheter ablation treatment for non-valvular atrial fibrillation (AF). Methods: We used a linked dataset including hospital administrative data, prescription medicine claims, emergency department presentations, and death registrations of patients in New South Wales, Australia. The cohort included patients who received catheter ablation for AF. Traditional and deep survival models were trained to predict major bleeding events and a composite of heart failure, stroke, cardiac arrest, and death. Results: Out of a total of 3285 patients in the cohort, 177 (5.3%) experienced the composite outcomeheart failure, stroke, cardiac arrest, deathand 167 (5.1%) experienced major bleeding events after catheter ablation treatment. Models predicting the composite outcome had high risk discrimination accuracy, with the best model having a concordance index > 0.79 at the evaluated time horizons. Models for predicting major bleeding events had poor risk discrimination performance, with all models having a concordance index < 0.66. The most impactful features for the models predicting higher risk were comorbidities indicative of poor health, older age, and therapies commonly used in sicker patients to treat heart failure and AF. Conclusions: Diagnosis and medication history did not contain sufficient information for precise risk prediction of experiencing major bleeding events. The models for predicting the composite outcome have the potential to enable clinicians to identify and manage high-risk patients following catheter ablation proactively. Future research is needed to validate the usefulness of these models in clinical practice.
Incorporating Uncertainty in Learning to Defer Algorithms for Safe Computer-Aided Diagnosis
Sep 03, 2021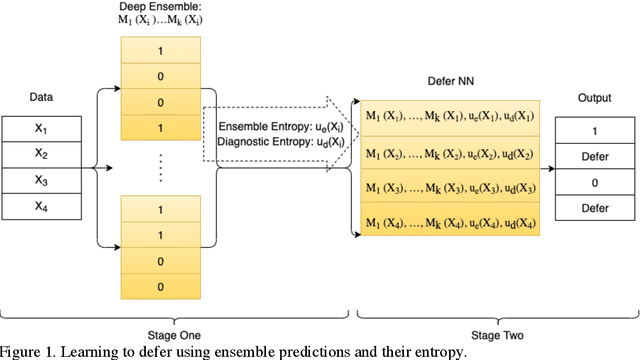
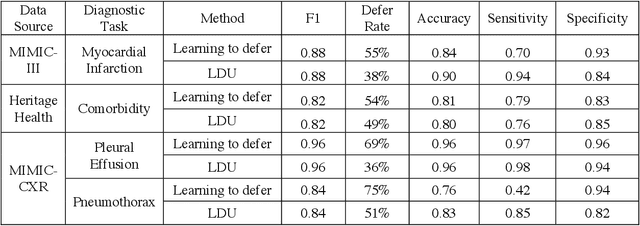
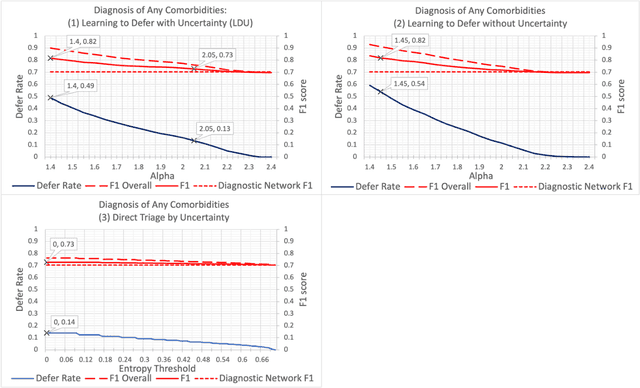
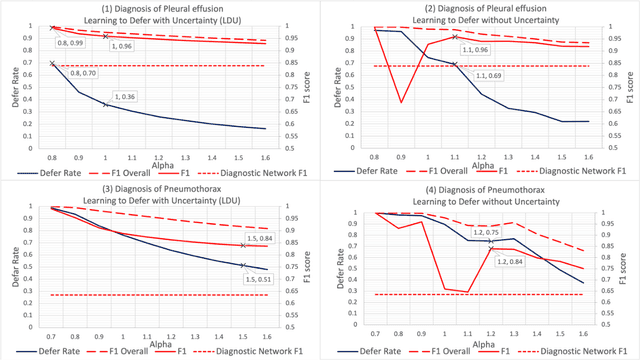
Abstract:In this study we propose the Learning to Defer with Uncertainty (LDU) algorithm, an approach which considers the model's predictive uncertainty when identifying the patient group to be evaluated by human experts. By identifying patients for whom the uncertainty of computer-aided diagnosis is estimated to be high and defers them for evaluation by human experts, the LDU algorithm can be used to mitigate the risk of erroneous computer-aided diagnoses in clinical settings.
Stochastic Treatment Recommendation with Deep Survival Dose Response Function (DeepSDRF)
Aug 24, 2021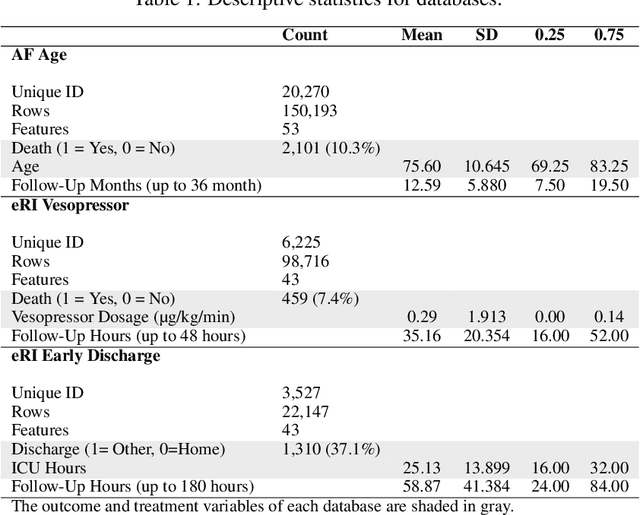
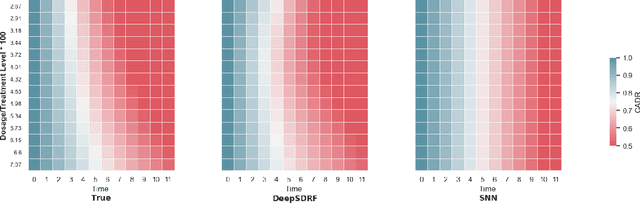
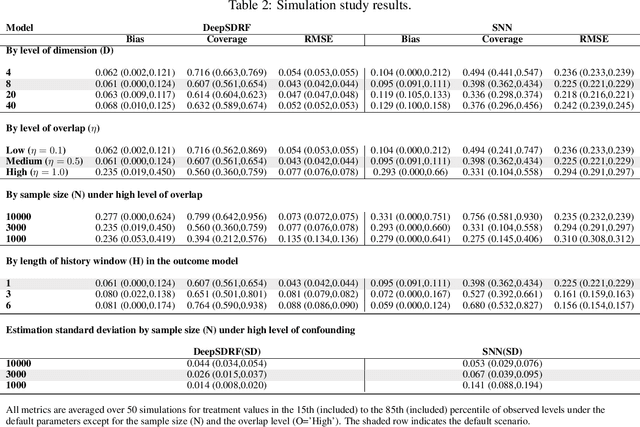
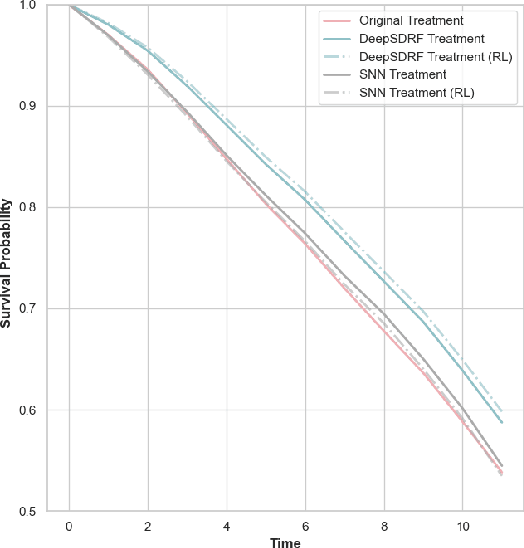
Abstract:We propose a general formulation for stochastic treatment recommendation problems in settings with clinical survival data, which we call the Deep Survival Dose Response Function (DeepSDRF). That is, we consider the problem of learning the conditional average dose response (CADR) function solely from historical data in which unobserved factors (confounders) affect both observed treatment and time-to-event outcomes. The estimated treatment effect from DeepSDRF enables us to develop recommender algorithms with explanatory insights. We compared two recommender approaches based on random search and reinforcement learning and found similar performance in terms of patient outcome. We tested the DeepSDRF and the corresponding recommender on extensive simulation studies and two empirical databases: 1) the Clinical Practice Research Datalink (CPRD) and 2) the eICU Research Institute (eRI) database. To the best of our knowledge, this is the first time that confounders are taken into consideration for addressing the stochastic treatment effect with observational data in a medical context.
CDSM -- Casual Inference using Deep Bayesian Dynamic Survival Models
Feb 24, 2021
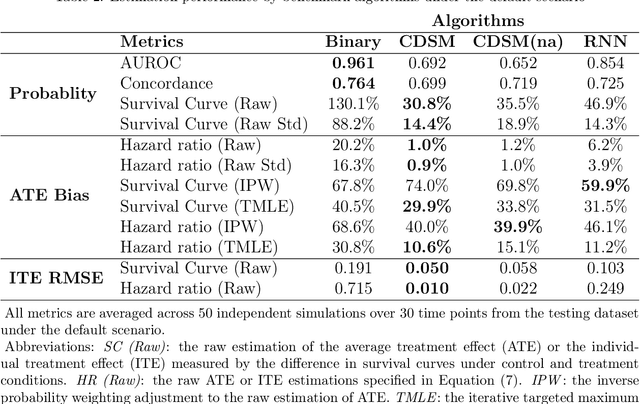
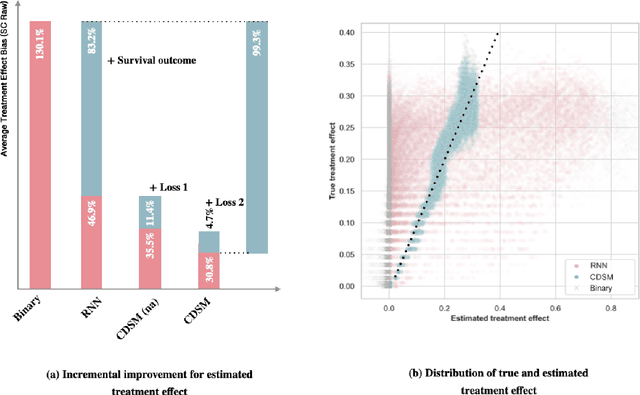
Abstract:A smart healthcare system that supports clinicians for risk-calibrated treatment assessment typically requires the accurate modeling of time-to-event outcomes. To tackle this sequential treatment effect estimation problem, we developed causal dynamic survival model (CDSM) for causal inference with survival outcomes using longitudinal electronic health record (EHR). CDSM has impressive explanatory performance while maintaining the prediction capability of conventional binary neural network predictors. It borrows the strength from explanatory framework including the survival analysis and counterfactual framework and integrates them with the prediction power from a deep Bayesian recurrent neural network to extract implicit knowledge from EHR data. In two large clinical cohort studies, our model identified the conditional average treatment effect in accordance with previous literature yet detected individual effect heterogeneity over time and patient subgroups. The model provides individualized and clinically interpretable treatment effect estimations to improve patient outcomes.
Dynamic prediction of time to event with survival curves
Jan 26, 2021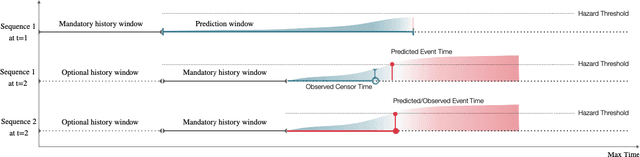
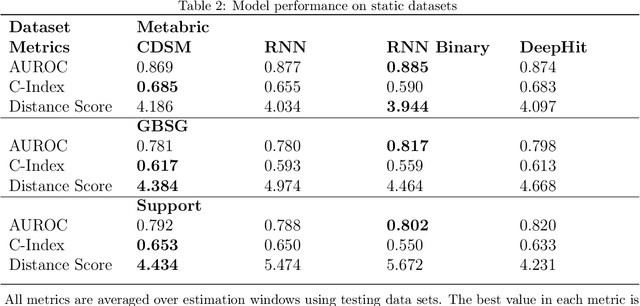
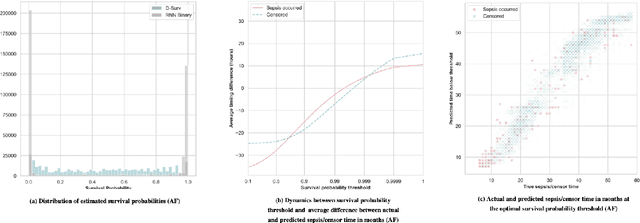
Abstract:With the ever-growing complexity of primary health care system, proactive patient failure management is an effective way to enhancing the availability of health care resource. One key enabler is the dynamic prediction of time-to-event outcomes. Conventional explanatory statistical approach lacks the capability of making precise individual level prediction, while the data adaptive binary predictors does not provide nominal survival curves for biologically plausible survival analysis. The purpose of this article is to elucidate that the knowledge of explanatory survival analysis can significantly enhance the current black-box data adaptive prediction models. We apply our recently developed counterfactual dynamic survival model (CDSM) to static and longitudinal observational data and testify that the inflection point of its estimated individual survival curves provides reliable prediction of the patient failure time.
Targeted Estimation of Heterogeneous Treatment Effect in Observational Survival Analysis
Oct 22, 2019
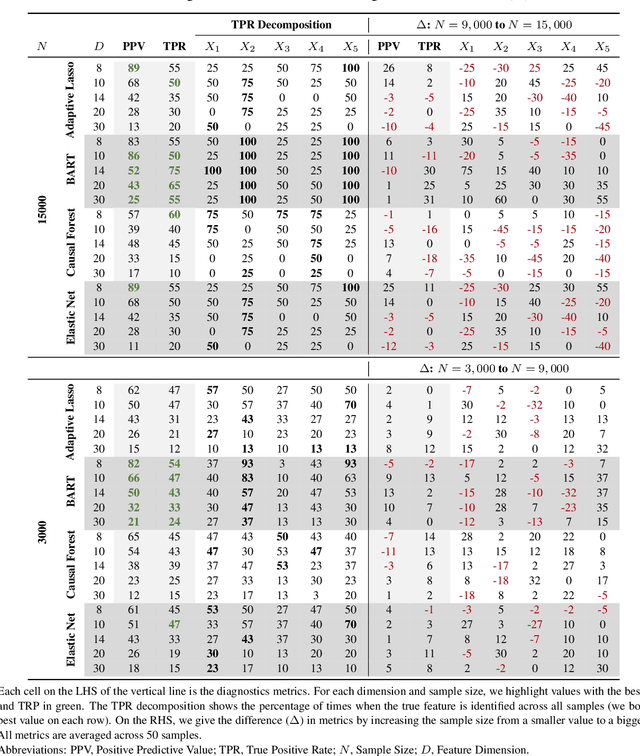
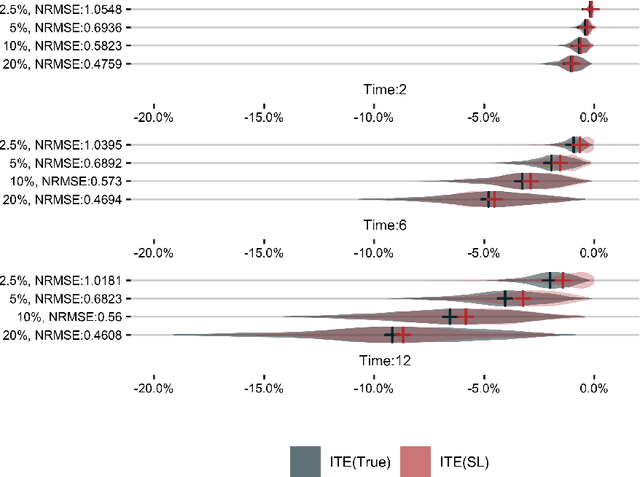
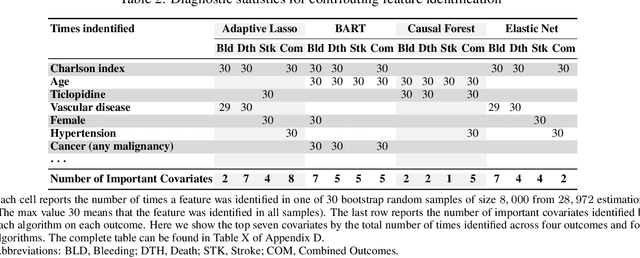
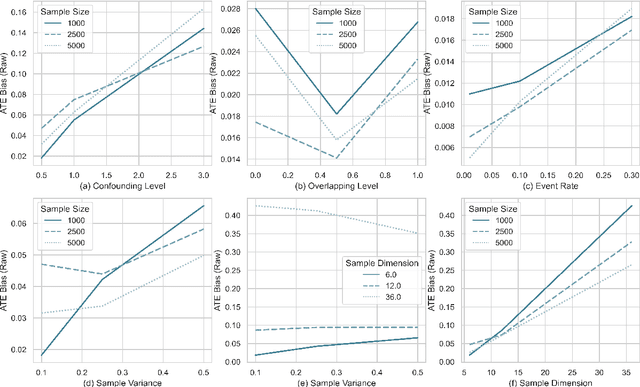
Abstract:The aim of clinical effectiveness research using repositories of electronic health records is to identify what health interventions 'work best' in real-world settings. Since there are several reasons why the net benefit of intervention may differ across patients, current comparative effectiveness literature focuses on investigating heterogeneous treatment effect and predicting whether an individual might benefit from an intervention. The majority of this literature has concentrated on the estimation of the effect of treatment on binary outcomes. However, many medical interventions are evaluated in terms of their effect on future events, which are subject to loss to follow-up. In this study, we describe a framework for the estimation of heterogeneous treatment effect in terms of differences in time-to-event (survival) probabilities. We divide the problem into three phases: (1) estimation of treatment effect conditioned on unique sets of the covariate vector; (2) identification of features important for heterogeneity using an ensemble of non-parametric variable importance methods; and (3) estimation of treatment effect on the reference classes defined by the previously selected features, using one-step Targeted Maximum Likelihood Estimation. We conducted a series of simulation studies and found that this method performs well when either sample size or event rate is high enough and the number of covariates contributing to the effect heterogeneity is moderate. An application of this method to a clinical case study was conducted by estimating the effect of oral anticoagulants on newly diagnosed non-valvular atrial fibrillation patients using data from the UK Clinical Practice Research Datalink.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge