Jinghui Liu
Rephrasing Electronic Health Records for Pretraining Clinical Language Models
Nov 28, 2024Abstract:Clinical language models are important for many applications in healthcare, but their development depends on access to extensive clinical text for pretraining. However, obtaining clinical notes from electronic health records (EHRs) at scale is challenging due to patient privacy concerns. In this study, we rephrase existing clinical notes using LLMs to generate synthetic pretraining corpora, drawing inspiration from previous work on rephrasing web data. We examine four popular small-sized LLMs (<10B) to create synthetic clinical text to pretrain both decoder-based and encoder-based language models. The method yields better results in language modeling and downstream tasks than previous synthesis approaches without referencing real clinical text. We find that augmenting original clinical notes with synthetic corpora from different LLMs improves performances even at a small token budget, showing the potential of this method to support pretraining at the institutional level or be scaled to synthesize large-scale clinical corpora.
e-Health CSIRO at RRG24: Entropy-Augmented Self-Critical Sequence Training for Radiology Report Generation
Aug 07, 2024Abstract:The Shared Task on Large-Scale Radiology Report Generation (RRG24) aims to expedite the development of assistive systems for interpreting and reporting on chest X-ray (CXR) images. This task challenges participants to develop models that generate the findings and impression sections of radiology reports from CXRs from a patient's study, using five different datasets. This paper outlines the e-Health CSIRO team's approach, which achieved multiple first-place finishes in RRG24. The core novelty of our approach lies in the addition of entropy regularisation to self-critical sequence training, to maintain a higher entropy in the token distribution. This prevents overfitting to common phrases and ensures a broader exploration of the vocabulary during training, essential for handling the diversity of the radiology reports in the RRG24 datasets. Our model is available on Hugging Face https://huggingface.co/aehrc/cxrmate-rrg24.
e-Health CSIRO at "Discharge Me!" 2024: Generating Discharge Summary Sections with Fine-tuned Language Models
Jul 03, 2024Abstract:Clinical documentation is an important aspect of clinicians' daily work and often demands a significant amount of time. The BioNLP 2024 Shared Task on Streamlining Discharge Documentation (Discharge Me!) aims to alleviate this documentation burden by automatically generating discharge summary sections, including brief hospital course and discharge instruction, which are often time-consuming to synthesize and write manually. We approach the generation task by fine-tuning multiple open-sourced language models (LMs), including both decoder-only and encoder-decoder LMs, with various configurations on input context. We also examine different setups for decoding algorithms, model ensembling or merging, and model specialization. Our results show that conditioning on the content of discharge summary prior to the target sections is effective for the generation task. Furthermore, we find that smaller encoder-decoder LMs can work as well or even slightly better than larger decoder based LMs fine-tuned through LoRA. The model checkpoints from our team (aehrc) are openly available.
Improving Text-based Early Prediction by Distillation from Privileged Time-Series Text
Jan 26, 2023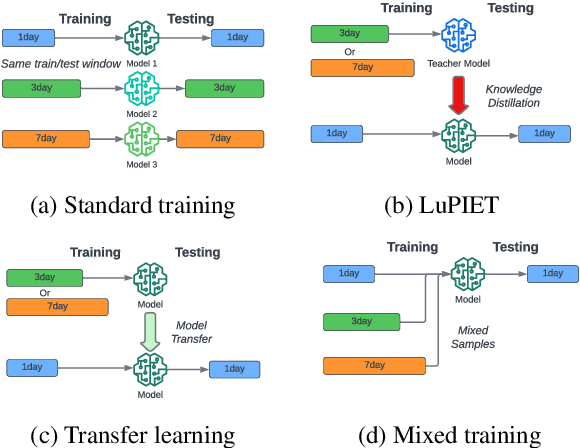
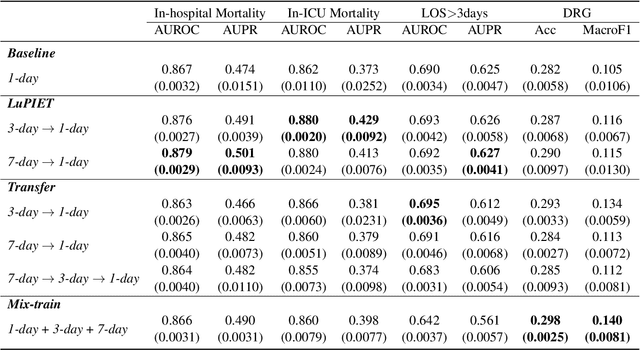
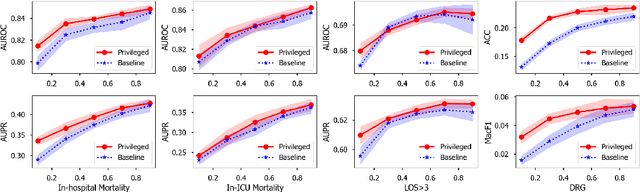
Abstract:Modeling text-based time-series to make prediction about a future event or outcome is an important task with a wide range of applications. The standard approach is to train and test the model using the same input window, but this approach neglects the data collected in longer input windows between the prediction time and the final outcome, which are often available during training. In this study, we propose to treat this neglected text as privileged information available during training to enhance early prediction modeling through knowledge distillation, presented as Learning using Privileged tIme-sEries Text (LuPIET). We evaluate the method on clinical and social media text, with four clinical prediction tasks based on clinical notes and two mental health prediction tasks based on social media posts. Our results show LuPIET is effective in enhancing text-based early predictions, though one may need to consider choosing the appropriate text representation and windows for privileged text to achieve optimal performance. Compared to two other methods using transfer learning and mixed training, LuPIET offers more stable improvements over the baseline, standard training. As far as we are concerned, this is the first study to examine learning using privileged information for time-series in the NLP context.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge