Olivier Salvado
NC-Reg : Neural Cortical Maps for Rigid Registration
Jan 26, 2026Abstract:We introduce neural cortical maps, a continuous and compact neural representation for cortical feature maps, as an alternative to traditional discrete structures such as grids and meshes. It can learn from meshes of arbitrary size and provide learnt features at any resolution. Neural cortical maps enable efficient optimization on the sphere and achieve runtimes up to 30 times faster than classic barycentric interpolation (for the same number of iterations). As a proof of concept, we investigate rigid registration of cortical surfaces and propose NC-Reg, a novel iterative algorithm that involves the use of neural cortical feature maps, gradient descent optimization and a simulated annealing strategy. Through ablation studies and subject-to-template experiments, our method demonstrates sub-degree accuracy ($<1^\circ$ from the global optimum), and serves as a promising robust pre-alignment strategy, which is critical in clinical settings.
Non-Invasive 3D Wound Measurement with RGB-D Imaging
Jan 26, 2026Abstract:Chronic wound monitoring and management require accurate and efficient wound measurement methods. This paper presents a fast, non-invasive 3D wound measurement algorithm based on RGB-D imaging. The method combines RGB-D odometry with B-spline surface reconstruction to generate detailed 3D wound meshes, enabling automatic computation of clinically relevant wound measurements such as perimeter, surface area, and dimensions. We evaluated our system on realistic silicone wound phantoms and measured sub-millimetre 3D reconstruction accuracy compared with high-resolution ground-truth scans. The extracted measurements demonstrated low variability across repeated captures and strong agreement with manual assessments. The proposed pipeline also outperformed a state-of-the-art object-centric RGB-D reconstruction method while maintaining runtimes suitable for real-time clinical deployment. Our approach offers a promising tool for automated wound assessment in both clinical and remote healthcare settings.
VAE with Hyperspherical Coordinates: Improving Anomaly Detection from Hypervolume-Compressed Latent Space
Jan 25, 2026Abstract:Variational autoencoders (VAE) encode data into lower-dimensional latent vectors before decoding those vectors back to data. Once trained, one can hope to detect out-of-distribution (abnormal) latent vectors, but several issues arise when the latent space is high dimensional. This includes an exponential growth of the hypervolume with the dimension, which severely affects the generative capacity of the VAE. In this paper, we draw insights from high dimensional statistics: in these regimes, the latent vectors of a standard VAE are distributed on the `equators' of a hypersphere, challenging the detection of anomalies. We propose to formulate the latent variables of a VAE using hyperspherical coordinates, which allows compressing the latent vectors towards a given direction on the hypersphere, thereby allowing for a more expressive approximate posterior. We show that this improves both the fully unsupervised and OOD anomaly detection ability of the VAE, achieving the best performance on the datasets we considered, outperforming existing methods. For the unsupervised and OOD modalities, respectively, these are: i) detecting unusual landscape from the Mars Rover camera and unusual Galaxies from ground based imagery (complex, real world datasets); ii) standard benchmarks like Cifar10 and subsets of ImageNet as the in-distribution (ID) class.
Multi-View Consistent Wound Segmentation With Neural Fields
Jan 23, 2026Abstract:Wound care is often challenged by the economic and logistical burdens that consistently afflict patients and hospitals worldwide. In recent decades, healthcare professionals have sought support from computer vision and machine learning algorithms. In particular, wound segmentation has gained interest due to its ability to provide professionals with fast, automatic tissue assessment from standard RGB images. Some approaches have extended segmentation to 3D, enabling more complete and precise healing progress tracking. However, inferring multi-view consistent 3D structures from 2D images remains a challenge. In this paper, we evaluate WoundNeRF, a NeRF SDF-based method for estimating robust wound segmentations from automatically generated annotations. We demonstrate the potential of this paradigm in recovering accurate segmentations by comparing it against state-of-the-art Vision Transformer networks and conventional rasterisation-based algorithms. The code will be released to facilitate further development in this promising paradigm.
Wound3DAssist: A Practical Framework for 3D Wound Assessment
Aug 25, 2025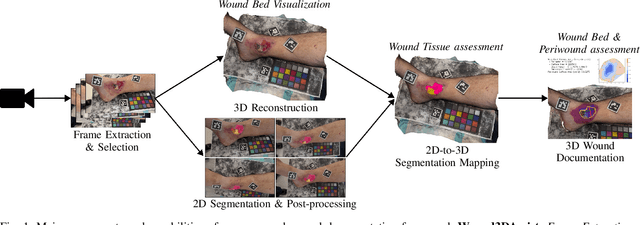
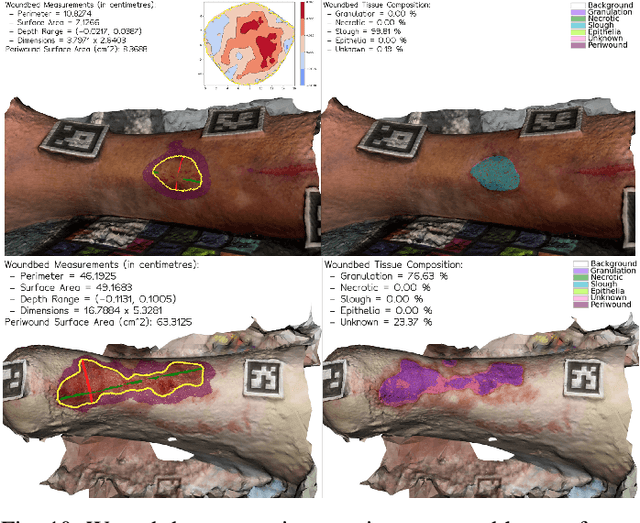
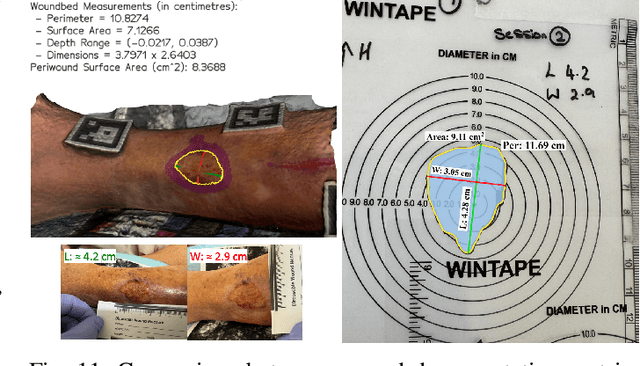
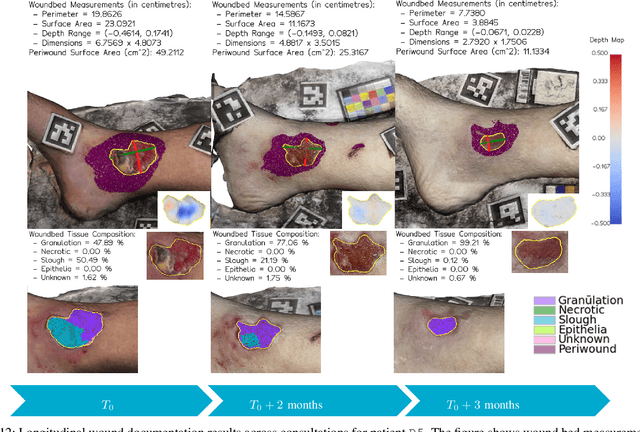
Abstract:Managing chronic wounds remains a major healthcare challenge, with clinical assessment often relying on subjective and time-consuming manual documentation methods. Although 2D digital videometry frameworks aided the measurement process, these approaches struggle with perspective distortion, a limited field of view, and an inability to capture wound depth, especially in anatomically complex or curved regions. To overcome these limitations, we present Wound3DAssist, a practical framework for 3D wound assessment using monocular consumer-grade videos. Our framework generates accurate 3D models from short handheld smartphone video recordings, enabling non-contact, automatic measurements that are view-independent and robust to camera motion. We integrate 3D reconstruction, wound segmentation, tissue classification, and periwound analysis into a modular workflow. We evaluate Wound3DAssist across digital models with known geometry, silicone phantoms, and real patients. Results show that the framework supports high-quality wound bed visualization, millimeter-level accuracy, and reliable tissue composition analysis. Full assessments are completed in under 20 minutes, demonstrating feasibility for real-world clinical use.
PINNs for Medical Image Analysis: A Survey
Aug 02, 2024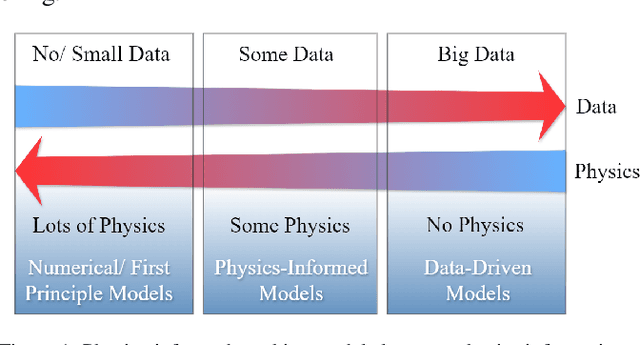

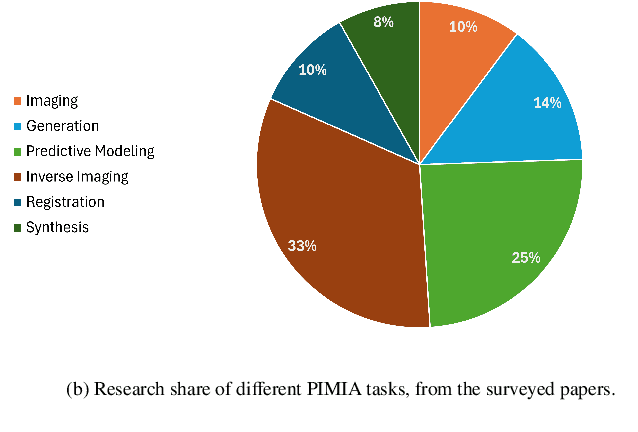

Abstract:The incorporation of physical information in machine learning frameworks is transforming medical image analysis (MIA). By integrating fundamental knowledge and governing physical laws, these models achieve enhanced robustness and interpretability. In this work, we explore the utility of physics-informed approaches for MIA (PIMIA) tasks such as registration, generation, classification, and reconstruction. We present a systematic literature review of over 80 papers on physics-informed methods dedicated to MIA. We propose a unified taxonomy to investigate what physics knowledge and processes are modelled, how they are represented, and the strategies to incorporate them into MIA models. We delve deep into a wide range of image analysis tasks, from imaging, generation, prediction, inverse imaging (super-resolution and reconstruction), registration, and image analysis (segmentation and classification). For each task, we thoroughly examine and present in a tabular format the central physics-guided operation, the region of interest (with respect to human anatomy), the corresponding imaging modality, the dataset used for model training, the deep network architecture employed, and the primary physical process, equation, or principle utilized. Additionally, we also introduce a novel metric to compare the performance of PIMIA methods across different tasks and datasets. Based on this review, we summarize and distil our perspectives on the challenges, open research questions, and directions for future research. We highlight key open challenges in PIMIA, including selecting suitable physics priors and establishing a standardized benchmarking platform.
SALVE: A 3D Reconstruction Benchmark of Wounds from Consumer-grade Videos
Jul 29, 2024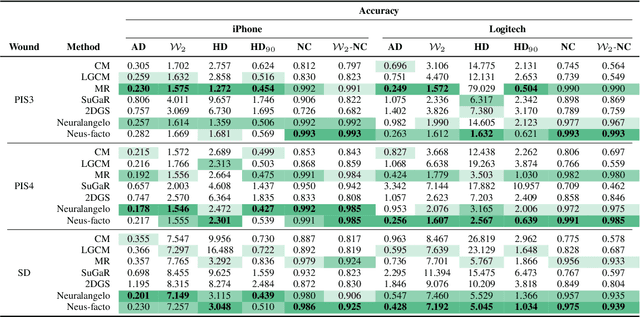
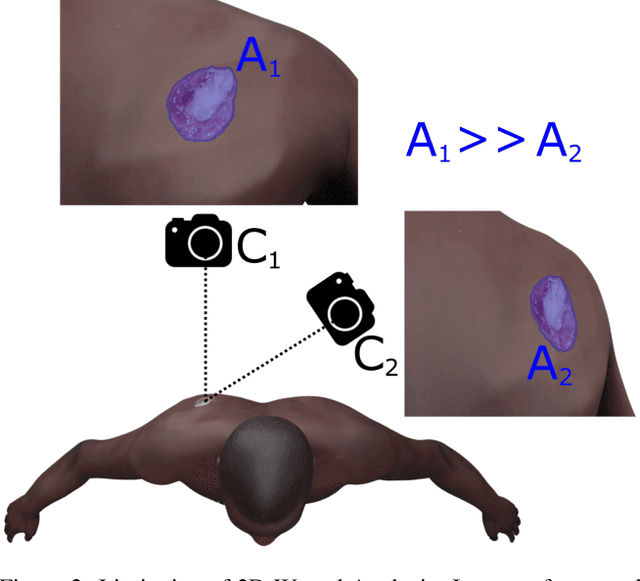
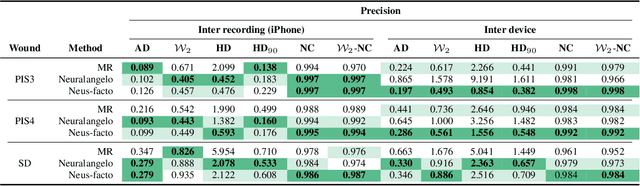
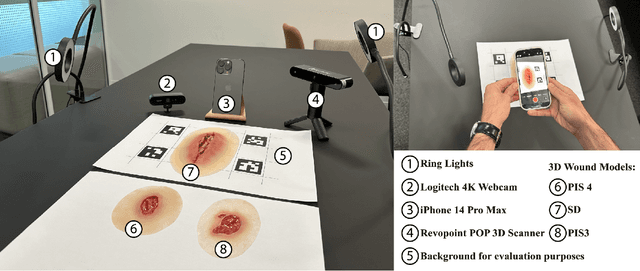
Abstract:Managing chronic wounds is a global challenge that can be alleviated by the adoption of automatic systems for clinical wound assessment from consumer-grade videos. While 2D image analysis approaches are insufficient for handling the 3D features of wounds, existing approaches utilizing 3D reconstruction methods have not been thoroughly evaluated. To address this gap, this paper presents a comprehensive study on 3D wound reconstruction from consumer-grade videos. Specifically, we introduce the SALVE dataset, comprising video recordings of realistic wound phantoms captured with different cameras. Using this dataset, we assess the accuracy and precision of state-of-the-art methods for 3D reconstruction, ranging from traditional photogrammetry pipelines to advanced neural rendering approaches. In our experiments, we observe that photogrammetry approaches do not provide smooth surfaces suitable for precise clinical measurements of wounds. Neural rendering approaches show promise in addressing this issue, advancing the use of this technology in wound care practices.
NeRF Director: Revisiting View Selection in Neural Volume Rendering
Jun 13, 2024Abstract:Neural Rendering representations have significantly contributed to the field of 3D computer vision. Given their potential, considerable efforts have been invested to improve their performance. Nonetheless, the essential question of selecting training views is yet to be thoroughly investigated. This key aspect plays a vital role in achieving high-quality results and aligns with the well-known tenet of deep learning: "garbage in, garbage out". In this paper, we first illustrate the importance of view selection by demonstrating how a simple rotation of the test views within the most pervasive NeRF dataset can lead to consequential shifts in the performance rankings of state-of-the-art techniques. To address this challenge, we introduce a unified framework for view selection methods and devise a thorough benchmark to assess its impact. Significant improvements can be achieved without leveraging error or uncertainty estimation but focusing on uniform view coverage of the reconstructed object, resulting in a training-free approach. Using this technique, we show that high-quality renderings can be achieved faster by using fewer views. We conduct extensive experiments on both synthetic datasets and realistic data to demonstrate the effectiveness of our proposed method compared with random, conventional error-based, and uncertainty-guided view selection.
Divide and Conquer: Rethinking the Training Paradigm of Neural Radiance Fields
Jan 29, 2024Abstract:Neural radiance fields (NeRFs) have exhibited potential in synthesizing high-fidelity views of 3D scenes but the standard training paradigm of NeRF presupposes an equal importance for each image in the training set. This assumption poses a significant challenge for rendering specific views presenting intricate geometries, thereby resulting in suboptimal performance. In this paper, we take a closer look at the implications of the current training paradigm and redesign this for more superior rendering quality by NeRFs. Dividing input views into multiple groups based on their visual similarities and training individual models on each of these groups enables each model to specialize on specific regions without sacrificing speed or efficiency. Subsequently, the knowledge of these specialized models is aggregated into a single entity via a teacher-student distillation paradigm, enabling spatial efficiency for online render-ing. Empirically, we evaluate our novel training framework on two publicly available datasets, namely NeRF synthetic and Tanks&Temples. Our evaluation demonstrates that our DaC training pipeline enhances the rendering quality of a state-of-the-art baseline model while exhibiting convergence to a superior minimum.
Syn3DWound: A Synthetic Dataset for 3D Wound Bed Analysis
Nov 27, 2023


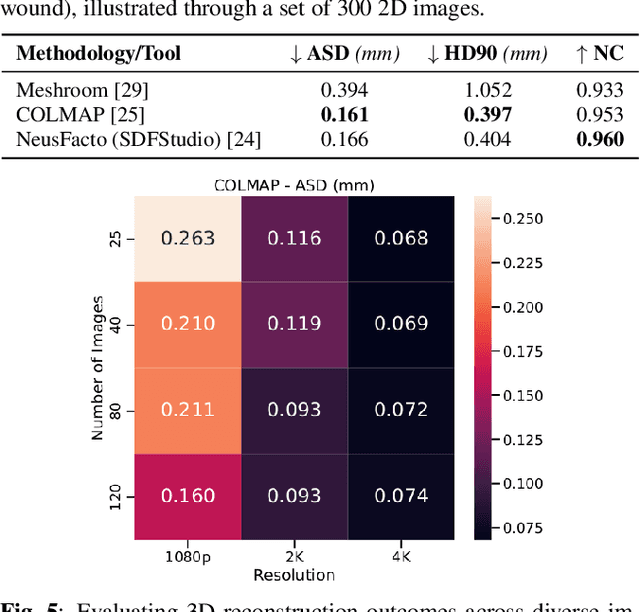
Abstract:Wound management poses a significant challenge, particularly for bedridden patients and the elderly. Accurate diagnostic and healing monitoring can significantly benefit from modern image analysis, providing accurate and precise measurements of wounds. Despite several existing techniques, the shortage of expansive and diverse training datasets remains a significant obstacle to constructing machine learning-based frameworks. This paper introduces Syn3DWound, an open-source dataset of high-fidelity simulated wounds with 2D and 3D annotations. We propose baseline methods and a benchmarking framework for automated 3D morphometry analysis and 2D/3D wound segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge