Fatima Nasrallah
Cascaded Multi-Modal Mixing Transformers for Alzheimer's Disease Classification with Incomplete Data
Oct 01, 2022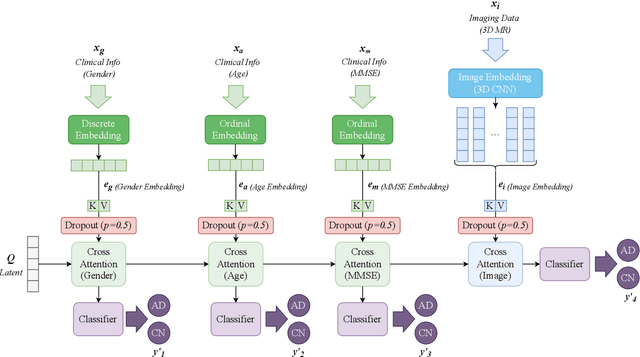
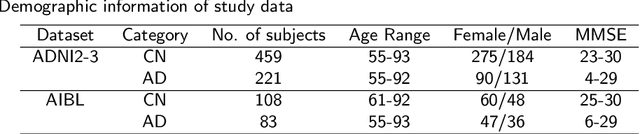
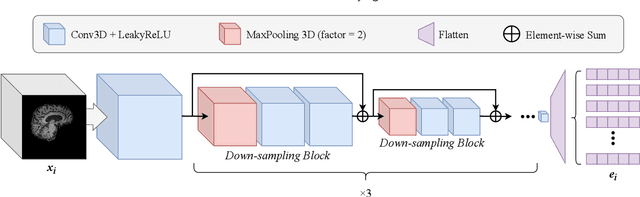
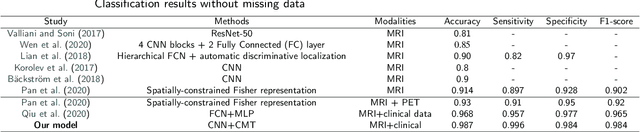
Abstract:Accurate medical classification requires a large number of multi-modal data, and in many cases, in different formats. Previous studies have shown promising results when using multi-modal data, outperforming single-modality models on when classifying disease such as AD. However, those models are usually not flexible enough to handle missing modalities. Currently, the most common workaround is excluding samples with missing modalities which leads to considerable data under-utilisation. Adding to the fact that labelled medical images are already scarce, the performance of data-driven methods like deep learning is severely hampered. Therefore, a multi-modal method that can gracefully handle missing data in various clinical settings is highly desirable. In this paper, we present the Multi-Modal Mixing Transformer (3MT), a novel Transformer for disease classification based on multi-modal data. In this work, we test it for \ac{AD} or \ac{CN} classification using neuroimaging data, gender, age and MMSE scores. The model uses a novel Cascaded Modality Transformers architecture with cross-attention to incorporate multi-modal information for more informed predictions. Auxiliary outputs and a novel modality dropout mechanism were incorporated to ensure an unprecedented level of modality independence and robustness. The result is a versatile network that enables the mixing of an unlimited number of modalities with different formats and full data utilization. 3MT was first tested on the ADNI dataset and achieved state-of-the-art test accuracy of $0.987\pm0.0006$. To test its generalisability, 3MT was directly applied to the AIBL after training on the ADNI dataset, and achieved a test accuracy of $0.925\pm0.0004$ without fine-tuning. Finally, we show that Grad-CAM visualizations are also possible with our model for explainable results.
Structure Guided Manifolds for Discovery of Disease Characteristics
Sep 24, 2022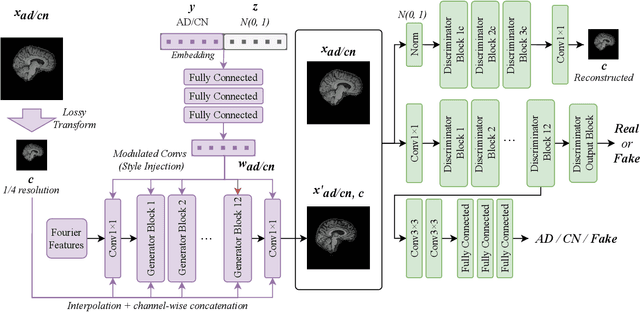
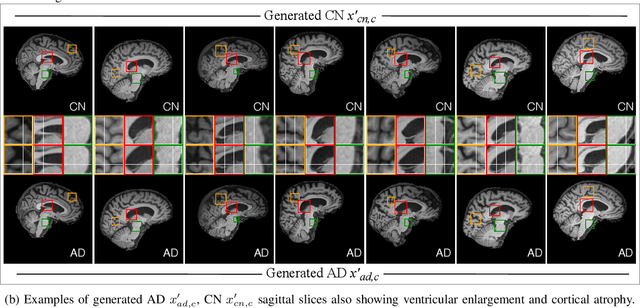
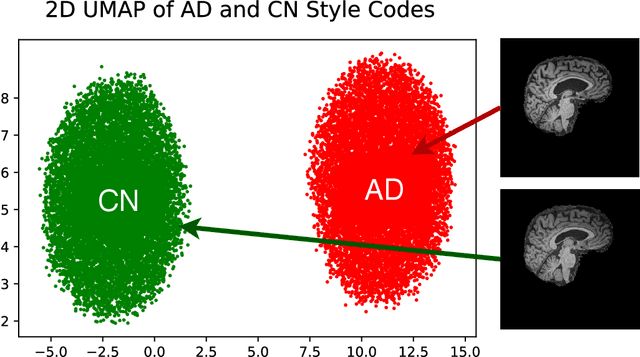

Abstract:In medical image analysis, the subtle visual characteristics of many diseases are challenging to discern, particularly due to the lack of paired data. For example, in mild Alzheimer's Disease (AD), brain tissue atrophy can be difficult to observe from pure imaging data, especially without paired AD and Cognitively Normal ( CN ) data for comparison. This work presents Disease Discovery GAN ( DiDiGAN), a weakly-supervised style-based framework for discovering and visualising subtle disease features. DiDiGAN learns a disease manifold of AD and CN visual characteristics, and the style codes sampled from this manifold are imposed onto an anatomical structural "blueprint" to synthesise paired AD and CN magnetic resonance images (MRIs). To suppress non-disease-related variations between the generated AD and CN pairs, DiDiGAN leverages a structural constraint with cycle consistency and anti-aliasing to enforce anatomical correspondence. When tested on the Alzheimer's Disease Neuroimaging Initiative ( ADNI) dataset, DiDiGAN showed key AD characteristics (reduced hippocampal volume, ventricular enlargement, and atrophy of cortical structures) through synthesising paired AD and CN scans. The qualitative results were backed up by automated brain volume analysis, where systematic pair-wise reductions in brain tissue structures were also measured
Instant tissue field and magnetic susceptibility mapping from MR raw phase using Laplacian enabled deep neural networks
Nov 16, 2021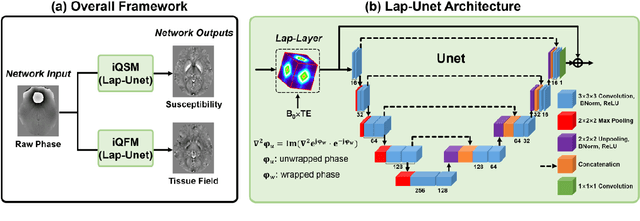

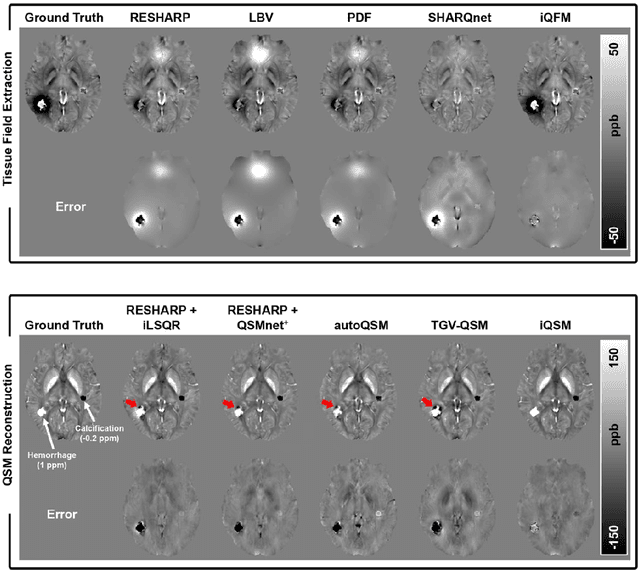

Abstract:Quantitative susceptibility mapping (QSM) is a valuable MRI post-processing technique that quantifies the magnetic susceptibility of body tissue from phase data. However, the traditional QSM reconstruction pipeline involves multiple non-trivial steps, including phase unwrapping, background field removal, and dipole inversion. These intermediate steps not only increase the reconstruction time but amplify noise and errors. This study develops a large-stencil Laplacian preprocessed deep learning-based neural network for near instant quantitative field and susceptibility mapping (i.e., iQFM and iQSM) from raw MR phase data. The proposed iQFM and iQSM methods were compared with established reconstruction pipelines on simulated and in vivo datasets. In addition, experiments on patients with intracranial hemorrhage and multiple sclerosis were also performed to test the generalization of the novel neural networks. The proposed iQFM and iQSM methods yielded comparable results to multi-step methods in healthy subjects while dramatically improving reconstruction accuracies on intracranial hemorrhages with large susceptibilities. The reconstruction time was also substantially shortened from minutes using multi-step methods to only 30 milliseconds using the trained iQFM and iQSM neural networks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge