Veronica Vilaplana
QU-BraTS: MICCAI BraTS 2020 Challenge on Quantifying Uncertainty in Brain Tumor Segmentation -- Analysis of Ranking Metrics and Benchmarking Results
Dec 19, 2021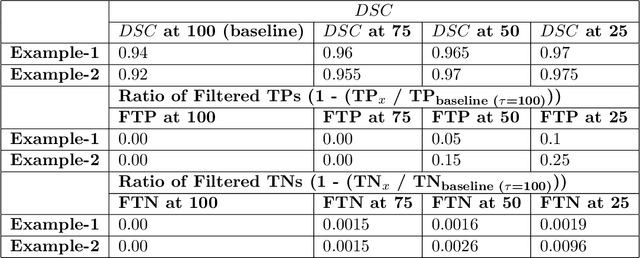
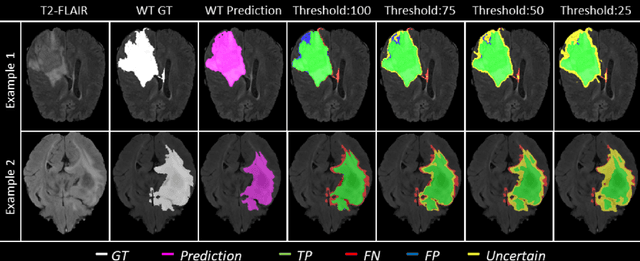
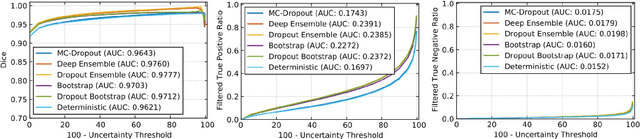
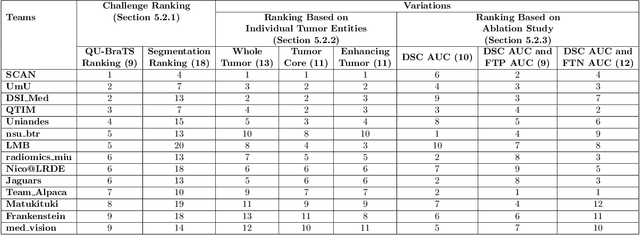
Abstract:Deep learning (DL) models have provided the state-of-the-art performance in a wide variety of medical imaging benchmarking challenges, including the Brain Tumor Segmentation (BraTS) challenges. However, the task of focal pathology multi-compartment segmentation (e.g., tumor and lesion sub-regions) is particularly challenging, and potential errors hinder the translation of DL models into clinical workflows. Quantifying the reliability of DL model predictions in the form of uncertainties, could enable clinical review of the most uncertain regions, thereby building trust and paving the way towards clinical translation. Recently, a number of uncertainty estimation methods have been introduced for DL medical image segmentation tasks. Developing metrics to evaluate and compare the performance of uncertainty measures will assist the end-user in making more informed decisions. In this study, we explore and evaluate a metric developed during the BraTS 2019-2020 task on uncertainty quantification (QU-BraTS), and designed to assess and rank uncertainty estimates for brain tumor multi-compartment segmentation. This metric (1) rewards uncertainty estimates that produce high confidence in correct assertions, and those that assign low confidence levels at incorrect assertions, and (2) penalizes uncertainty measures that lead to a higher percentages of under-confident correct assertions. We further benchmark the segmentation uncertainties generated by 14 independent participating teams of QU-BraTS 2020, all of which also participated in the main BraTS segmentation task. Overall, our findings confirm the importance and complementary value that uncertainty estimates provide to segmentation algorithms, and hence highlight the need for uncertainty quantification in medical image analyses. Our evaluation code is made publicly available at https://github.com/RagMeh11/QU-BraTS.
MRI brain tumor segmentation and uncertainty estimation using 3D-UNet architectures
Dec 30, 2020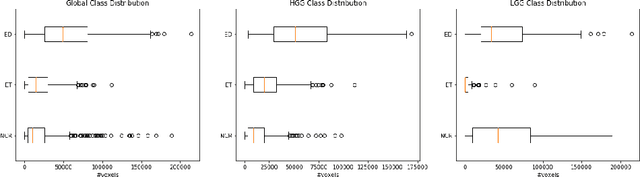
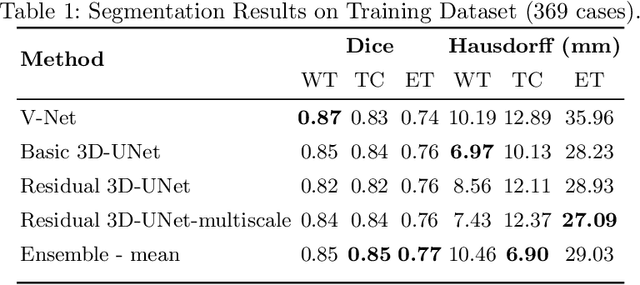
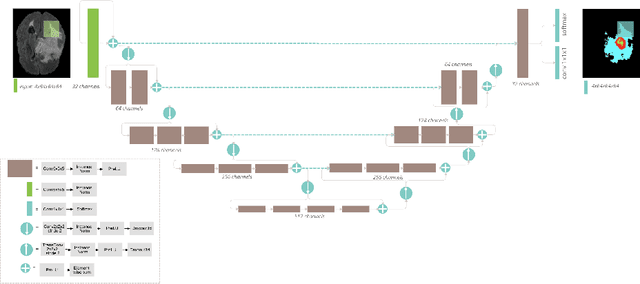
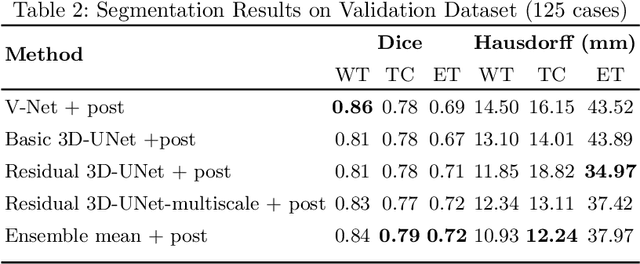
Abstract:Automation of brain tumor segmentation in 3D magnetic resonance images (MRIs) is key to assess the diagnostic and treatment of the disease. In recent years, convolutional neural networks (CNNs) have shown improved results in the task. However, high memory consumption is still a problem in 3D-CNNs. Moreover, most methods do not include uncertainty information, which is especially critical in medical diagnosis. This work studies 3D encoder-decoder architectures trained with patch-based techniques to reduce memory consumption and decrease the effect of unbalanced data. The different trained models are then used to create an ensemble that leverages the properties of each model, thus increasing the performance. We also introduce voxel-wise uncertainty information, both epistemic and aleatoric using test-time dropout (TTD) and data-augmentation (TTA) respectively. In addition, a hybrid approach is proposed that helps increase the accuracy of the segmentation. The model and uncertainty estimation measurements proposed in this work have been used in the BraTS'20 Challenge for task 1 and 3 regarding tumor segmentation and uncertainty estimation.
Brain Tumor Segmentation using 3D-CNNs with Uncertainty Estimation
Sep 24, 2020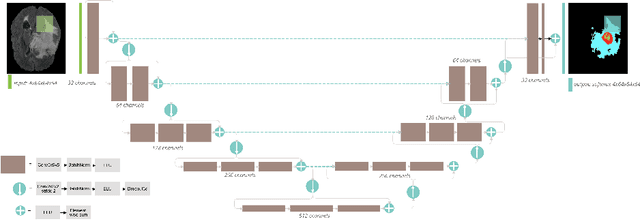
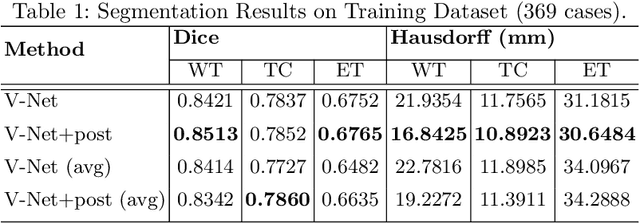
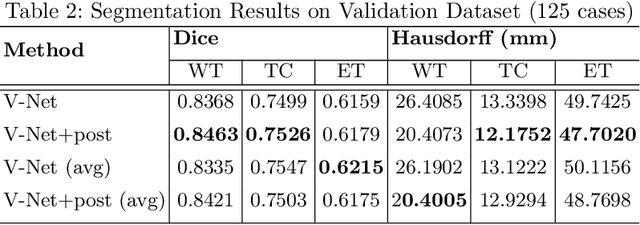
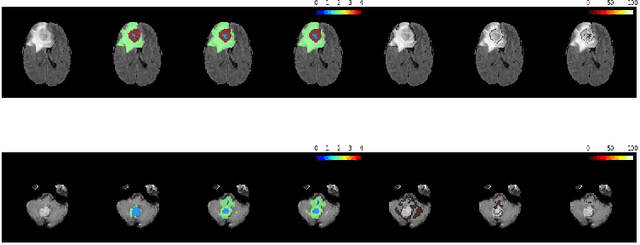
Abstract:Automation of brain tumors in 3D magnetic resonance images (MRIs) is key to assess the diagnostic and treatment of the disease. In recent years, convolutional neural networks (CNNs) have shown improved results in the task. However, high memory consumption is still a problem in 3D-CNNs. Moreover, most methods do not include uncertainty information, which is specially critical in medical diagnosis. This work proposes a 3D encoder-decoder architecture, based on V-Net \cite{vnet} which is trained with patching techniques to reduce memory consumption and decrease the effect of unbalanced data. We also introduce voxel-wise uncertainty, both epistemic and aleatoric using test-time dropout and data-augmentation respectively. Uncertainty maps can provide extra information to expert neurologists, useful for detecting when the model is not confident on the provided segmentation.
Picking groups instead of samples: A close look at Static Pool-based Meta-Active Learning
Nov 01, 2019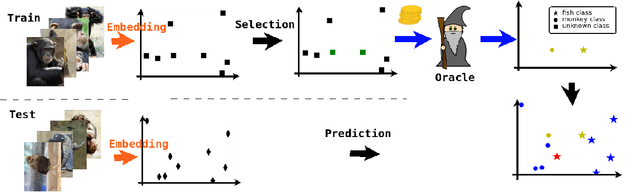
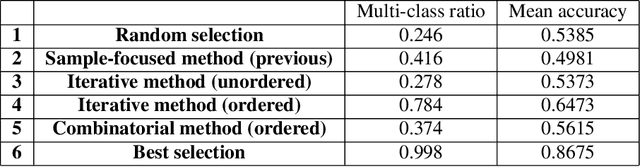
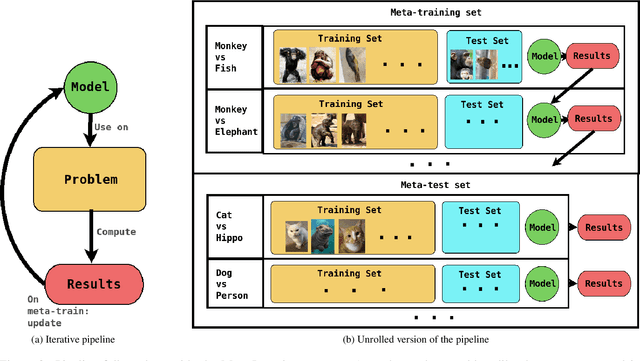
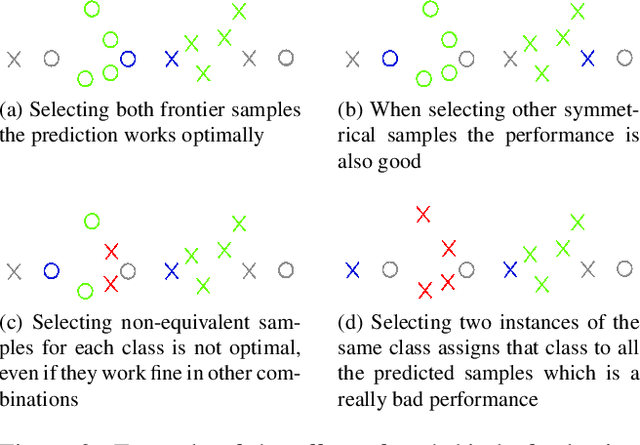
Abstract:Active Learning techniques are used to tackle learning problems where obtaining training labels is costly. In this work we use Meta-Active Learning to learn to select a subset of samples from a pool of unsupervised input for further annotation. This scenario is called Static Pool-based Meta- Active Learning. We propose to extend existing approaches by performing the selection in a manner that, unlike previous works, can handle the selection of each sample based on the whole selected subset.
BCN20000: Dermoscopic Lesions in the Wild
Aug 30, 2019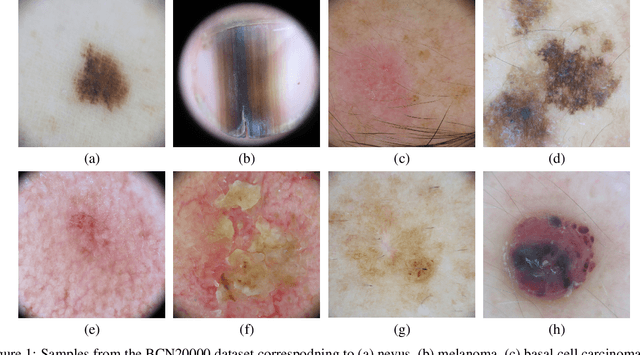
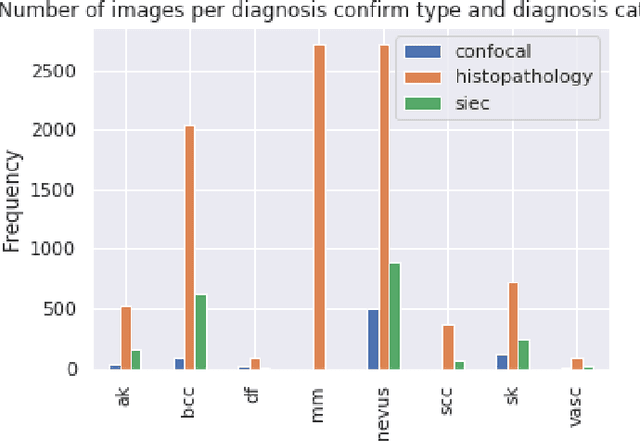
Abstract:This article summarizes the BCN20000 dataset, composed of 19424 dermoscopic images of skin lesions captured from 2010 to 2016 in the facilities of the Hospital Cl\'inic in Barcelona. With this dataset, we aim to study the problem of unconstrained classification of dermoscopic images of skin cancer, including lesions found in hard-to-diagnose locations (nails and mucosa), large lesions which do not fit in the aperture of the dermoscopy device, and hypo-pigmented lesions. The BCN20000 will be provided to the participants of the ISIC Challenge 2019, where they will be asked to train algorithms to classify dermoscopic images of skin cancer automatically.
Monte-Carlo Sampling applied to Multiple Instance Learning for Histological Image Classification
Dec 30, 2018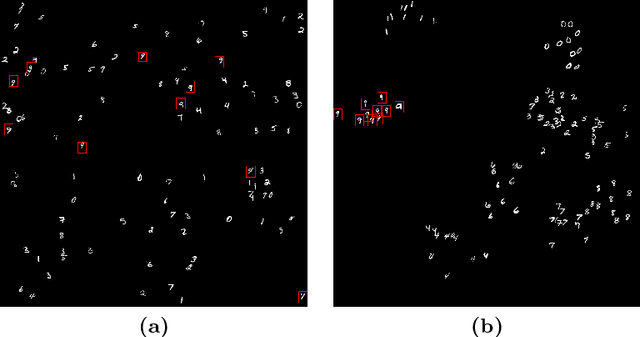
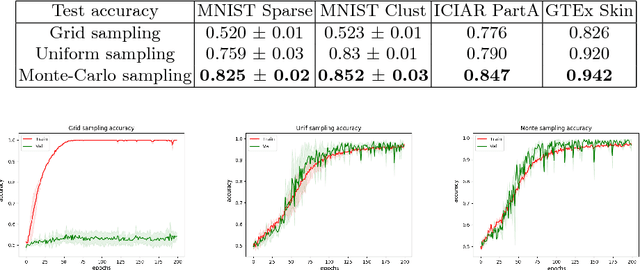
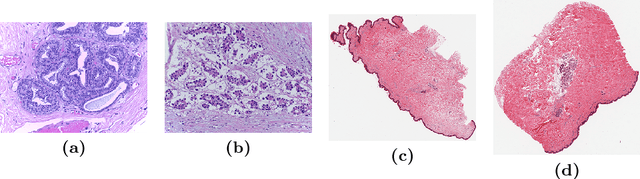
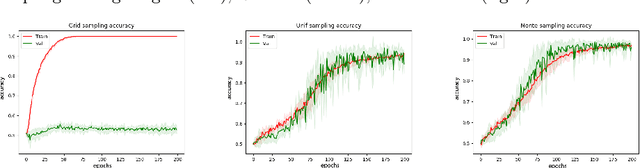
Abstract:We propose a patch sampling strategy based on a sequential Monte-Carlo method for high resolution image classification in the context of Multiple Instance Learning. When compared with grid sampling and uniform sampling techniques, it achieves higher generalization performance. We validate the strategy on two artificial datasets and two histological datasets for breast cancer and sun exposure classification.
Brain MRI super-resolution using 3D generative adversarial networks
Dec 29, 2018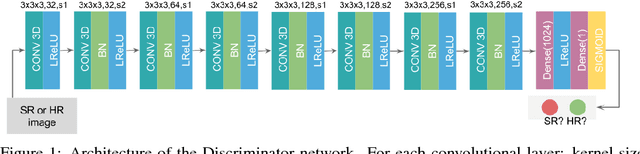
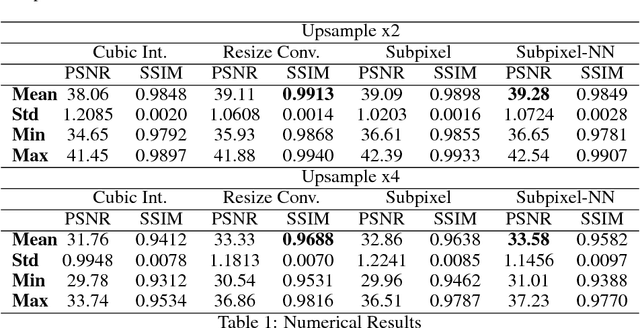
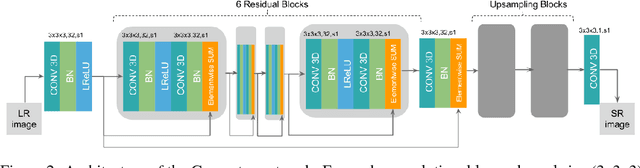
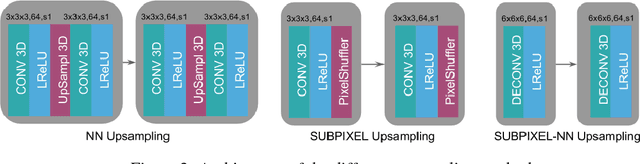
Abstract:In this work we propose an adversarial learning approach to generate high resolution MRI scans from low resolution images. The architecture, based on the SRGAN model, adopts 3D convolutions to exploit volumetric information. For the discriminator, the adversarial loss uses least squares in order to stabilize the training. For the generator, the loss function is a combination of a least squares adversarial loss and a content term based on mean square error and image gradients in order to improve the quality of the generated images. We explore different solutions for the upsampling phase. We present promising results that improve classical interpolation, showing the potential of the approach for 3D medical imaging super-resolution. Source code available at https://github.com/imatge-upc/3D-GAN-superresolution
Voxelwise nonlinear regression toolbox for neuroimage analysis: Application to aging and neurodegenerative disease modeling
Apr 18, 2017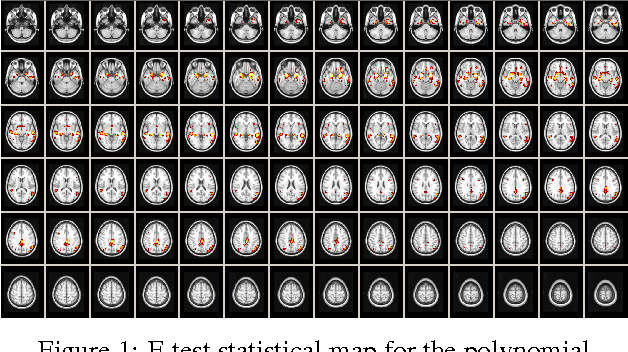
Abstract:This paper describes a new neuroimaging analysis toolbox that allows for the modeling of nonlinear effects at the voxel level, overcoming limitations of methods based on linear models like the GLM. We illustrate its features using a relevant example in which distinct nonlinear trajectories of Alzheimer's disease related brain atrophy patterns were found across the full biological spectrum of the disease. The open-source toolbox presented in this paper is available at https://github.com/imatge-upc/VNeAT.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge