Adrià Casamitjana
USLR: an open-source tool for unbiased and smooth longitudinal registration of brain MR
Nov 14, 2023Abstract:We present USLR, a computational framework for longitudinal registration of brain MRI scans to estimate nonlinear image trajectories that are smooth across time, unbiased to any timepoint, and robust to imaging artefacts. It operates on the Lie algebra parameterisation of spatial transforms (which is compatible with rigid transforms and stationary velocity fields for nonlinear deformation) and takes advantage of log-domain properties to solve the problem using Bayesian inference. USRL estimates rigid and nonlinear registrations that: (i) bring all timepoints to an unbiased subject-specific space; and (i) compute a smooth trajectory across the imaging time-series. We capitalise on learning-based registration algorithms and closed-form expressions for fast inference. A use-case Alzheimer's disease study is used to showcase the benefits of the pipeline in multiple fronts, such as time-consistent image segmentation to reduce intra-subject variability, subject-specific prediction or population analysis using tensor-based morphometry. We demonstrate that such approach improves upon cross-sectional methods in identifying group differences, which can be helpful in detecting more subtle atrophy levels or in reducing sample sizes in clinical trials. The code is publicly available in https://github.com/acasamitjana/uslr
Synth-by-Reg (SbR): Contrastive learning for synthesis-based registration of paired images
Jul 30, 2021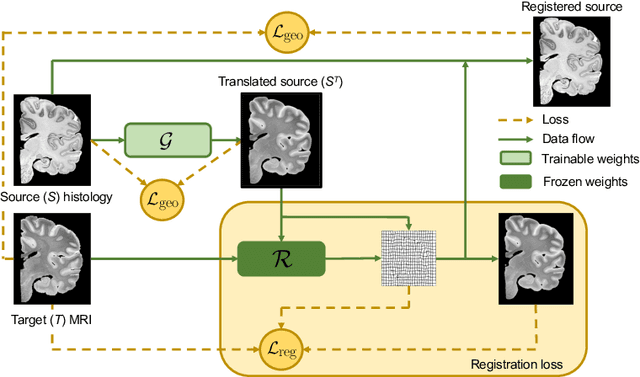


Abstract:Nonlinear inter-modality registration is often challenging due to the lack of objective functions that are good proxies for alignment. Here we propose a synthesis-by-registration method to convert this problem into an easier intra-modality task. We introduce a registration loss for weakly supervised image translation between domains that does not require perfectly aligned training data. This loss capitalises on a registration U-Net with frozen weights, to drive a synthesis CNN towards the desired translation. We complement this loss with a structure preserving constraint based on contrastive learning, which prevents blurring and content shifts due to overfitting. We apply this method to the registration of histological sections to MRI slices, a key step in 3D histology reconstruction. Results on two different public datasets show improvements over registration based on mutual information (13% reduction in landmark error) and synthesis-based algorithms such as CycleGAN (11% reduction), and are comparable to a registration CNN with label supervision.
Robust joint registration of multiple stains and MRI for multimodal 3D histology reconstruction: Application to the Allen human brain atlas
May 04, 2021
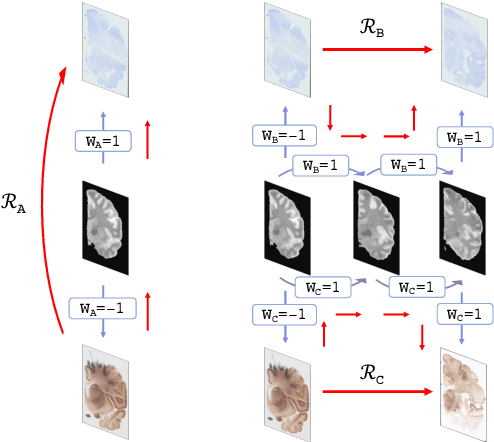


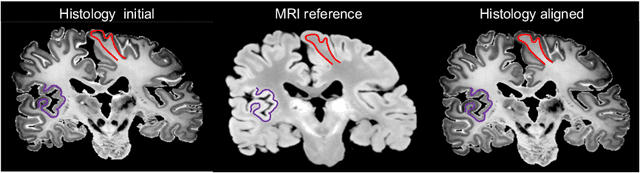
Abstract:Joint registration of a stack of 2D histological sections to recover 3D structure (3D histology reconstruction) finds application in areas such as atlas building and validation of in vivo imaging. Straighforward pairwise registration of neighbouring sections yields smooth reconstructions but has well-known problems such as banana effect (straightening of curved structures) and z-shift (drift). While these problems can be alleviated with an external, linearly aligned reference (e.g., Magnetic Resonance images), registration is often inaccurate due to contrast differences and the strong nonlinear distortion of the tissue, including artefacts such as folds and tears. In this paper, we present a probabilistic model of spatial deformation that yields reconstructions for multiple histological stains that that are jointly smooth, robust to outliers, and follow the reference shape. The model relies on a spanning tree of latent transforms connecting all the sections and slices, and assumes that the registration between any pair of images can be see as a noisy version of the composition of (possibly inverted) latent transforms connecting the two images. Bayesian inference is used to compute the most likely latent transforms given a set of pairwise registrations between image pairs within and across modalities. Results on synthetic deformations on multiple MR modalities, show that our method can accurately and robustly register multiple contrasts even in the presence of outliers. The 3D histology reconstruction of two stains (Nissl and parvalbumin) from the Allen human brain atlas, show its benefits on real data with severe distortions. We also provide the correspondence to MNI space, bridging the gap between two of the most used atlases in histology and MRI. Data is available at https://openneuro.org/datasets/ds003590 and code at https://github.com/acasamitjana/3dhirest.
DeepReg: a deep learning toolkit for medical image registration
Nov 04, 2020Abstract:DeepReg (https://github.com/DeepRegNet/DeepReg) is a community-supported open-source toolkit for research and education in medical image registration using deep learning.
Standardized Assessment of Automatic Segmentation of White Matter Hyperintensities and Results of the WMH Segmentation Challenge
Apr 01, 2019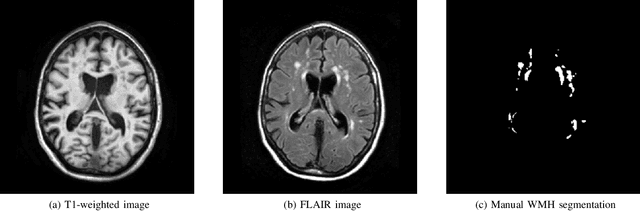
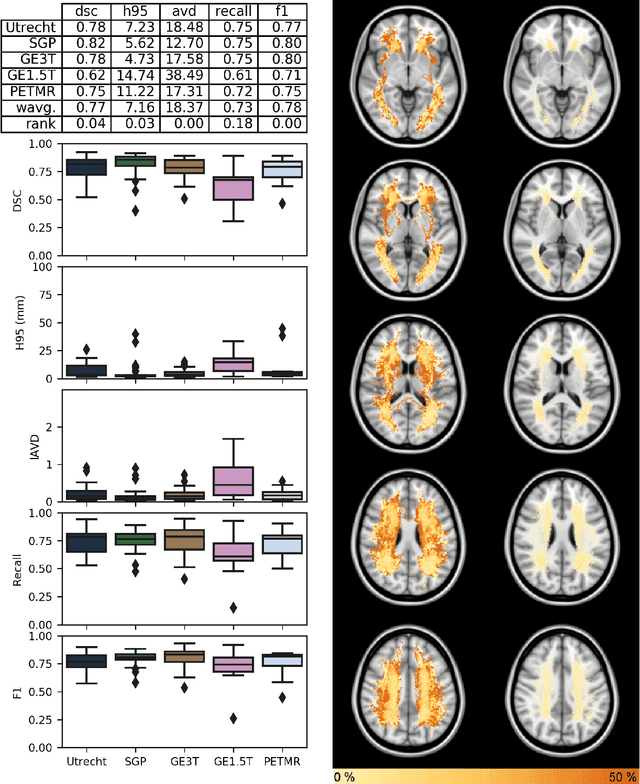
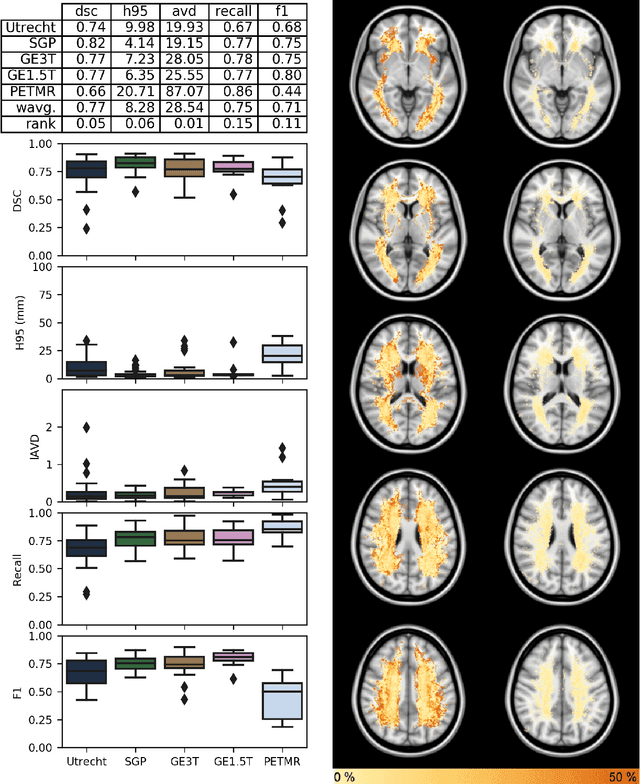
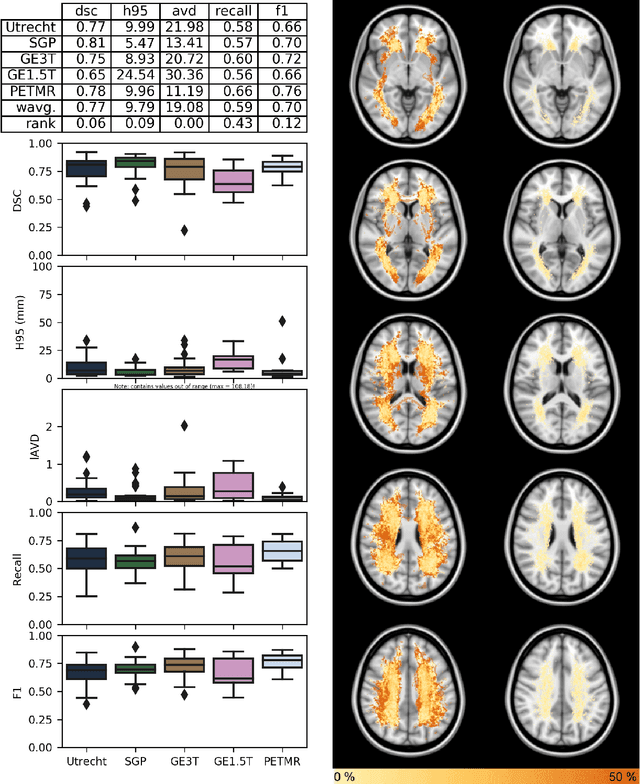
Abstract:Quantification of cerebral white matter hyperintensities (WMH) of presumed vascular origin is of key importance in many neurological research studies. Currently, measurements are often still obtained from manual segmentations on brain MR images, which is a laborious procedure. Automatic WMH segmentation methods exist, but a standardized comparison of the performance of such methods is lacking. We organized a scientific challenge, in which developers could evaluate their method on a standardized multi-center/-scanner image dataset, giving an objective comparison: the WMH Segmentation Challenge (https://wmh.isi.uu.nl/). Sixty T1+FLAIR images from three MR scanners were released with manual WMH segmentations for training. A test set of 110 images from five MR scanners was used for evaluation. Segmentation methods had to be containerized and submitted to the challenge organizers. Five evaluation metrics were used to rank the methods: (1) Dice similarity coefficient, (2) modified Hausdorff distance (95th percentile), (3) absolute log-transformed volume difference, (4) sensitivity for detecting individual lesions, and (5) F1-score for individual lesions. Additionally, methods were ranked on their inter-scanner robustness. Twenty participants submitted their method for evaluation. This paper provides a detailed analysis of the results. In brief, there is a cluster of four methods that rank significantly better than the other methods, with one clear winner. The inter-scanner robustness ranking shows that not all methods generalize to unseen scanners. The challenge remains open for future submissions and provides a public platform for method evaluation.
Cascaded V-Net using ROI masks for brain tumor segmentation
Dec 30, 2018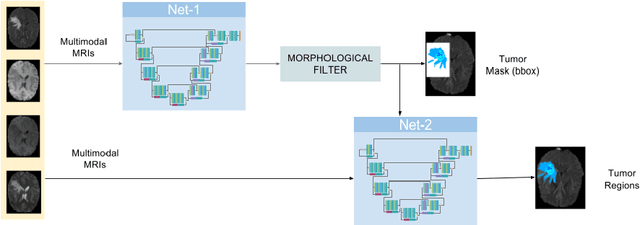
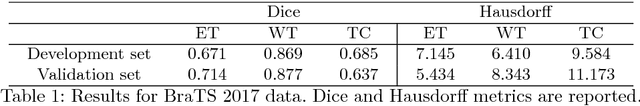
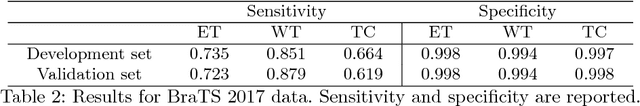
Abstract:In this work we approach the brain tumor segmentation problem with a cascade of two CNNs inspired in the V-Net architecture \cite{VNet}, reformulating residual connections and making use of ROI masks to constrain the networks to train only on relevant voxels. This architecture allows dense training on problems with highly skewed class distributions, such as brain tumor segmentation, by focusing training only on the vecinity of the tumor area. We report results on BraTS2017 Training and Validation sets.
* Third International Workshop, BrainLes 2017, Held in Conjunction with MICCAI 2017, Quebec City, QC, Canada, September 14, 2017, Revised Selected Papers
3D Convolutional Neural Networks for Brain Tumor Segmentation: A Comparison of Multi-resolution Architectures
May 23, 2017

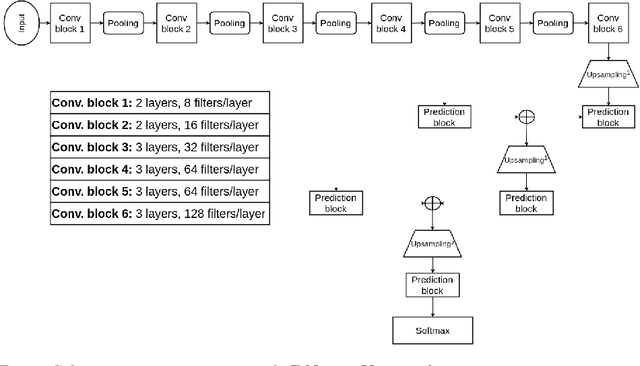

Abstract:This paper analyzes the use of 3D Convolutional Neural Networks for brain tumor segmentation in MR images. We address the problem using three different architectures that combine fine and coarse features to obtain the final segmentation. We compare three different networks that use multi-resolution features in terms of both design and performance and we show that they improve their single-resolution counterparts.
Voxelwise nonlinear regression toolbox for neuroimage analysis: Application to aging and neurodegenerative disease modeling
Apr 18, 2017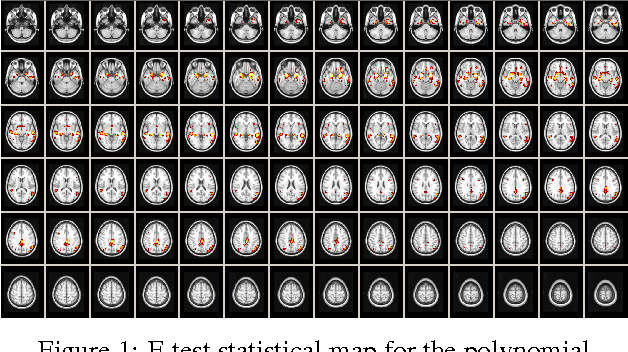
Abstract:This paper describes a new neuroimaging analysis toolbox that allows for the modeling of nonlinear effects at the voxel level, overcoming limitations of methods based on linear models like the GLM. We illustrate its features using a relevant example in which distinct nonlinear trajectories of Alzheimer's disease related brain atrophy patterns were found across the full biological spectrum of the disease. The open-source toolbox presented in this paper is available at https://github.com/imatge-upc/VNeAT.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge