Yunguan Fu
Semi-weakly-supervised neural network training for medical image registration
Feb 16, 2024



Abstract:For training registration networks, weak supervision from segmented corresponding regions-of-interest (ROIs) have been proven effective for (a) supplementing unsupervised methods, and (b) being used independently in registration tasks in which unsupervised losses are unavailable or ineffective. This correspondence-informing supervision entails cost in annotation that requires significant specialised effort. This paper describes a semi-weakly-supervised registration pipeline that improves the model performance, when only a small corresponding-ROI-labelled dataset is available, by exploiting unlabelled image pairs. We examine two types of augmentation methods by perturbation on network weights and image resampling, such that consistency-based unsupervised losses can be applied on unlabelled data. The novel WarpDDF and RegCut approaches are proposed to allow commutative perturbation between an image pair and the predicted spatial transformation (i.e. respective input and output of registration networks), distinct from existing perturbation methods for classification or segmentation. Experiments using 589 male pelvic MR images, labelled with eight anatomical ROIs, show the improvement in registration performance and the ablated contributions from the individual strategies. Furthermore, this study attempts to construct one of the first computational atlases for pelvic structures, enabled by registering inter-subject MRs, and quantifies the significant differences due to the proposed semi-weak supervision with a discussion on the potential clinical use of example atlas-derived statistics.
A Recycling Training Strategy for Medical Image Segmentation with Diffusion Denoising Models
Aug 30, 2023



Abstract:Denoising diffusion models have found applications in image segmentation by generating segmented masks conditioned on images. Existing studies predominantly focus on adjusting model architecture or improving inference such as test-time sampling strategies. In this work, we focus on training strategy improvements and propose a novel recycling method. During each training step, a segmentation mask is first predicted given an image and a random noise. This predicted mask, replacing the conventional ground truth mask, is used for denoising task during training. This approach can be interpreted as aligning the training strategy with inference by eliminating the dependence on ground truth masks for generating noisy samples. Our proposed method significantly outperforms standard diffusion training, self-conditioning, and existing recycling strategies across multiple medical imaging data sets: muscle ultrasound, abdominal CT, prostate MR, and brain MR. This holds true for two widely adopted sampling strategies: denoising diffusion probabilistic model and denoising diffusion implicit model. Importantly, existing diffusion models often display a declining or unstable performance during inference, whereas our novel recycling consistently enhances or maintains performance. Furthermore, we show for the first time that, under a fair comparison with the same network architectures and computing budget, the proposed recycling-based diffusion models achieved on-par performance with non-diffusion-based supervised training. This paper summarises these quantitative results and discusses their values, with a fully reproducible JAX-based implementation, released at https://github.com/mathpluscode/ImgX-DiffSeg.
Spatial Correspondence between Graph Neural Network-Segmented Images
Mar 17, 2023Abstract:Graph neural networks (GNNs) have been proposed for medical image segmentation, by predicting anatomical structures represented by graphs of vertices and edges. One such type of graph is predefined with fixed size and connectivity to represent a reference of anatomical regions of interest, thus known as templates. This work explores the potentials in these GNNs with common topology for establishing spatial correspondence, implicitly maintained during segmenting two or more images. With an example application of registering local vertebral sub-regions found in CT images, our experimental results showed that the GNN-based segmentation is capable of accurate and reliable localization of the same interventionally interesting structures between images, not limited to the segmentation classes. The reported average target registration errors of 2.2$\pm$1.3 mm and 2.7$\pm$1.4 mm, for aligning holdout test images with a reference and for aligning two test images, respectively, were by a considerable margin lower than those from the tested non-learning and learning-based registration algorithms. Further ablation studies assess the contributions towards the registration performance, from individual components in the originally segmentation-purposed network and its training algorithm. The results highlight that the proposed segmentation-in-lieu-of-registration approach shares methodological similarities with existing registration methods, such as the use of displacement smoothness constraint and point distance minimization albeit on non-grid graphs, which interestingly yielded benefits for both segmentation and registration. We, therefore, conclude that the template-based GNN segmentation can effectively establish spatial correspondence in our application, without any other dedicated registration algorithms.
Importance of Aligning Training Strategy with Evaluation for Diffusion Models in 3D Multiclass Segmentation
Mar 10, 2023Abstract:Recently, denoising diffusion probabilistic models (DDPM) have been applied to image segmentation by generating segmentation masks conditioned on images, while the applications were mainly limited to 2D networks without exploiting potential benefits from the 3D formulation. In this work, for the first time, DDPMs are used for 3D multiclass image segmentation. We make three key contributions that all focus on aligning the training strategy with the evaluation methodology, and improving efficiency. Firstly, the model predicts segmentation masks instead of sampled noise and is optimised directly via Dice loss. Secondly, the predicted mask in the previous time step is recycled to generate noise-corrupted masks to reduce information leakage. Finally, the diffusion process during training was reduced to five steps, the same as the evaluation. Through studies on two large multiclass data sets (prostate MR and abdominal CT), we demonstrated significantly improved performance compared to existing DDPMs, and reached competitive performance with non-diffusion segmentation models, based on U-net, within the same compute budget. The JAX-based diffusion framework has been released on https://github.com/mathpluscode/ImgX-DiffSeg.
Active learning using adaptable task-based prioritisation
Dec 03, 2022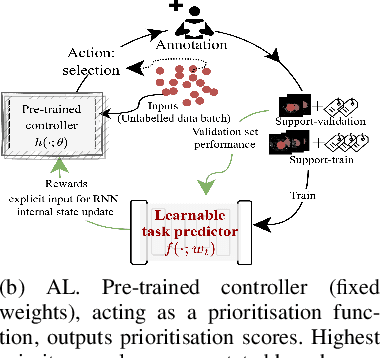
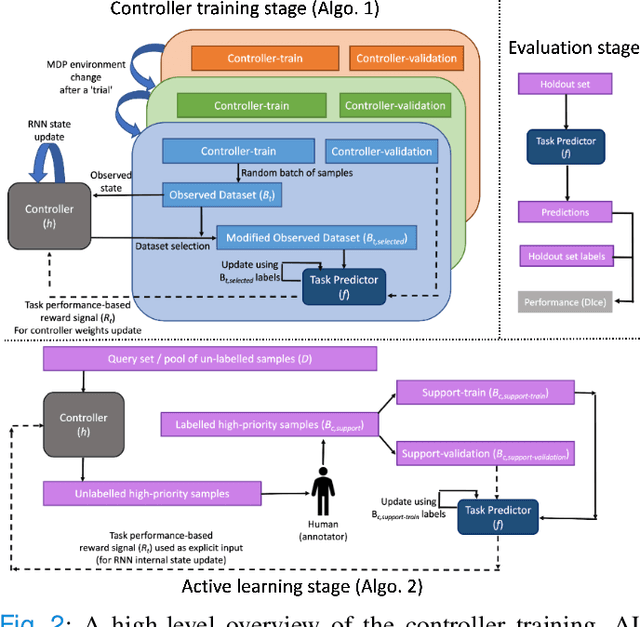
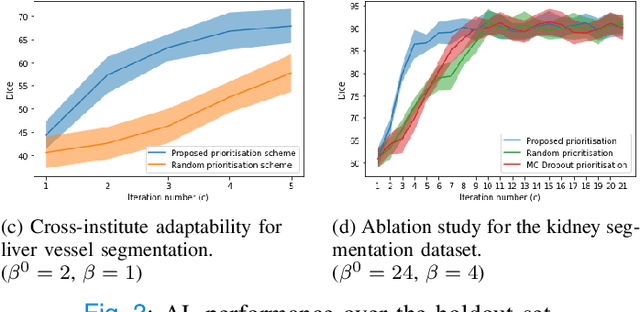
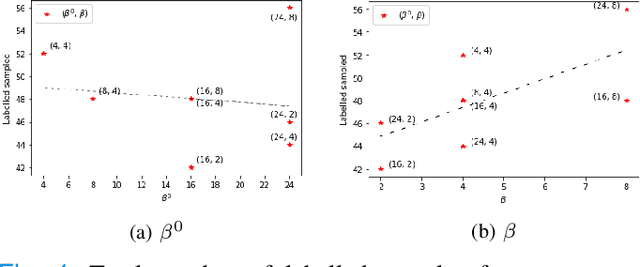
Abstract:Supervised machine learning-based medical image computing applications necessitate expert label curation, while unlabelled image data might be relatively abundant. Active learning methods aim to prioritise a subset of available image data for expert annotation, for label-efficient model training. We develop a controller neural network that measures priority of images in a sequence of batches, as in batch-mode active learning, for multi-class segmentation tasks. The controller is optimised by rewarding positive task-specific performance gain, within a Markov decision process (MDP) environment that also optimises the task predictor. In this work, the task predictor is a segmentation network. A meta-reinforcement learning algorithm is proposed with multiple MDPs, such that the pre-trained controller can be adapted to a new MDP that contains data from different institutes and/or requires segmentation of different organs or structures within the abdomen. We present experimental results using multiple CT datasets from more than one thousand patients, with segmentation tasks of nine different abdominal organs, to demonstrate the efficacy of the learnt prioritisation controller function and its cross-institute and cross-organ adaptability. We show that the proposed adaptable prioritisation metric yields converging segmentation accuracy for the novel class of kidney, unseen in training, using between approximately 40\% to 60\% of labels otherwise required with other heuristic or random prioritisation metrics. For clinical datasets of limited size, the proposed adaptable prioritisation offers a performance improvement of 22.6\% and 10.2\% in Dice score, for tasks of kidney and liver vessel segmentation, respectively, compared to random prioritisation and alternative active sampling strategies.
MONAI: An open-source framework for deep learning in healthcare
Nov 04, 2022



Abstract:Artificial Intelligence (AI) is having a tremendous impact across most areas of science. Applications of AI in healthcare have the potential to improve our ability to detect, diagnose, prognose, and intervene on human disease. For AI models to be used clinically, they need to be made safe, reproducible and robust, and the underlying software framework must be aware of the particularities (e.g. geometry, physiology, physics) of medical data being processed. This work introduces MONAI, a freely available, community-supported, and consortium-led PyTorch-based framework for deep learning in healthcare. MONAI extends PyTorch to support medical data, with a particular focus on imaging, and provide purpose-specific AI model architectures, transformations and utilities that streamline the development and deployment of medical AI models. MONAI follows best practices for software-development, providing an easy-to-use, robust, well-documented, and well-tested software framework. MONAI preserves the simple, additive, and compositional approach of its underlying PyTorch libraries. MONAI is being used by and receiving contributions from research, clinical and industrial teams from around the world, who are pursuing applications spanning nearly every aspect of healthcare.
Prototypical few-shot segmentation for cross-institution male pelvic structures with spatial registration
Sep 13, 2022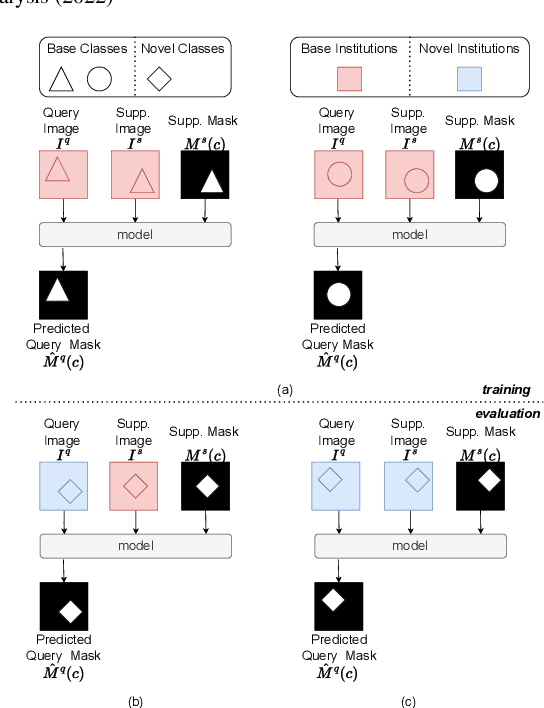

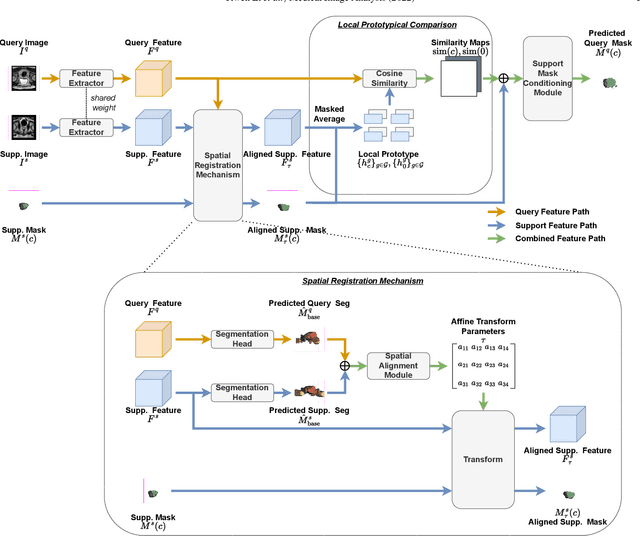
Abstract:The prowess that makes few-shot learning desirable in medical image analysis is the efficient use of the support image data, which are labelled to classify or segment new classes, a task that otherwise requires substantially more training images and expert annotations. This work describes a fully 3D prototypical few-shot segmentation algorithm, such that the trained networks can be effectively adapted to clinically interesting structures that are absent in training, using only a few labelled images from a different institute. First, to compensate for the widely recognised spatial variability between institutions in episodic adaptation of novel classes, a novel spatial registration mechanism is integrated into prototypical learning, consisting of a segmentation head and an spatial alignment module. Second, to assist the training with observed imperfect alignment, support mask conditioning module is proposed to further utilise the annotation available from the support images. Extensive experiments are presented in an application of segmenting eight anatomical structures important for interventional planning, using a data set of 589 pelvic T2-weighted MR images, acquired at seven institutes. The results demonstrate the efficacy in each of the 3D formulation, the spatial registration, and the support mask conditioning, all of which made positive contributions independently or collectively. Compared with the previously proposed 2D alternatives, the few-shot segmentation performance was improved with statistical significance, regardless whether the support data come from the same or different institutes.
Cross-Modality Image Registration using a Training-Time Privileged Third Modality
Jul 26, 2022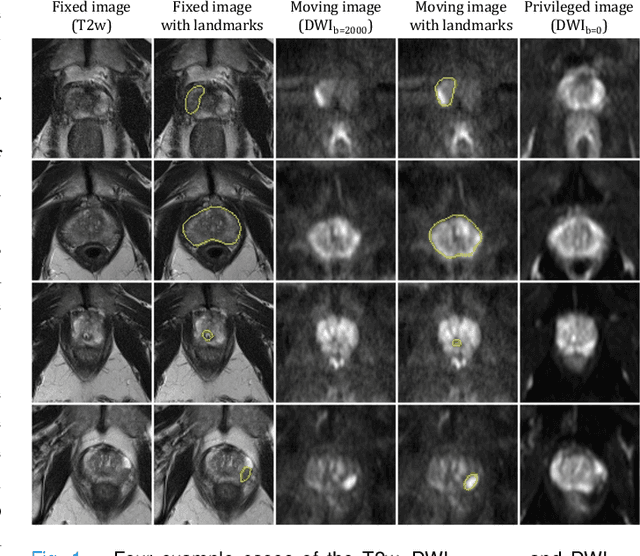
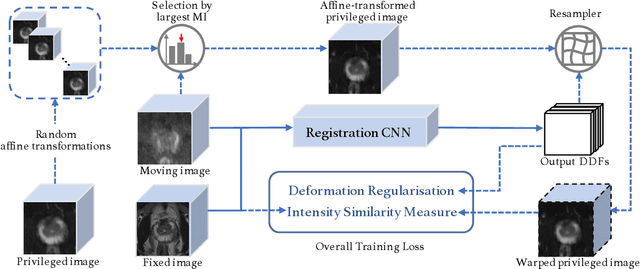
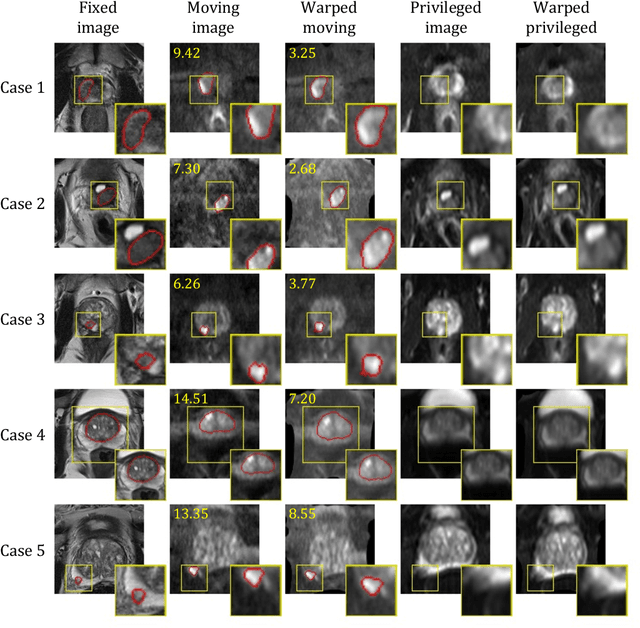
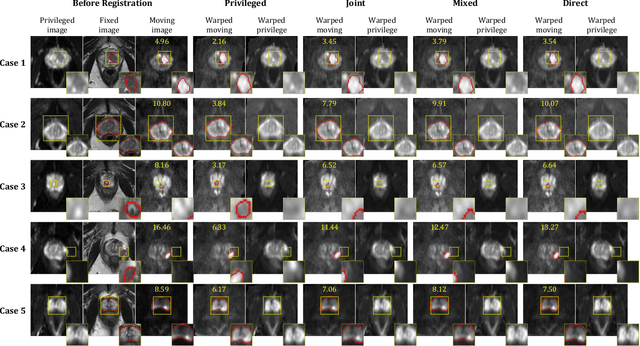
Abstract:In this work, we consider the task of pairwise cross-modality image registration, which may benefit from exploiting additional images available only at training time from an additional modality that is different to those being registered. As an example, we focus on aligning intra-subject multiparametric Magnetic Resonance (mpMR) images, between T2-weighted (T2w) scans and diffusion-weighted scans with high b-value (DWI$_{high-b}$). For the application of localising tumours in mpMR images, diffusion scans with zero b-value (DWI$_{b=0}$) are considered easier to register to T2w due to the availability of corresponding features. We propose a learning from privileged modality algorithm, using a training-only imaging modality DWI$_{b=0}$, to support the challenging multi-modality registration problems. We present experimental results based on 369 sets of 3D multiparametric MRI images from 356 prostate cancer patients and report, with statistical significance, a lowered median target registration error of 4.34 mm, when registering the holdout DWI$_{high-b}$ and T2w image pairs, compared with that of 7.96 mm before registration. Results also show that the proposed learning-based registration networks enabled efficient registration with comparable or better accuracy, compared with a classical iterative algorithm and other tested learning-based methods with/without the additional modality. These compared algorithms also failed to produce any significantly improved alignment between DWI$_{high-b}$ and T2w in this challenging application.
Image quality assessment for machine learning tasks using meta-reinforcement learning
Mar 27, 2022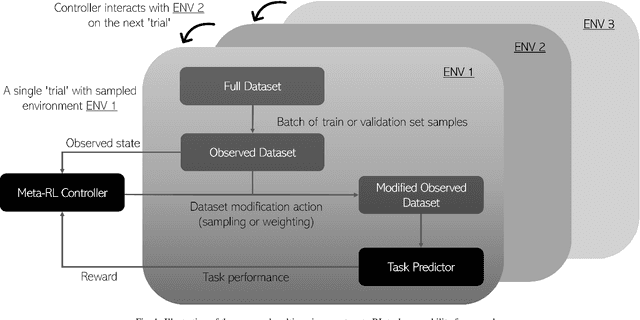
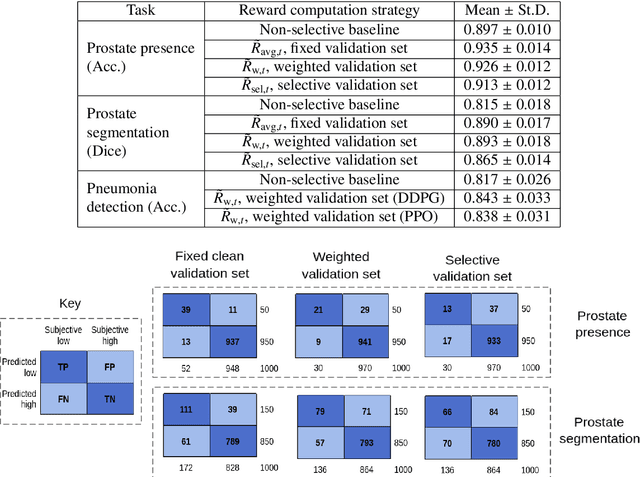
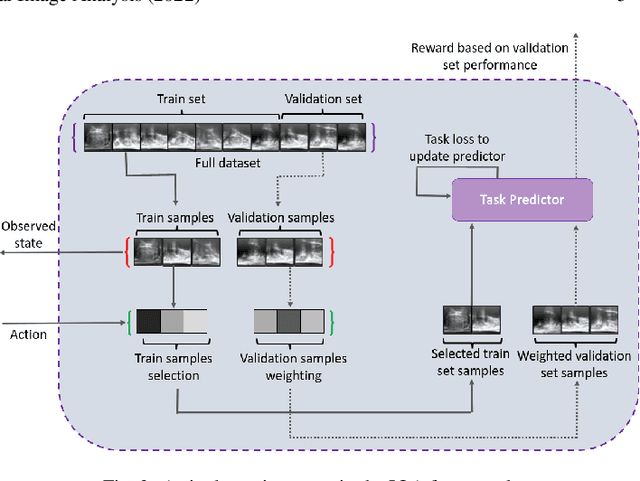
Abstract:In this paper, we consider image quality assessment (IQA) as a measure of how images are amenable with respect to a given downstream task, or task amenability. When the task is performed using machine learning algorithms, such as a neural-network-based task predictor for image classification or segmentation, the performance of the task predictor provides an objective estimate of task amenability. In this work, we use an IQA controller to predict the task amenability which, itself being parameterised by neural networks, can be trained simultaneously with the task predictor. We further develop a meta-reinforcement learning framework to improve the adaptability for both IQA controllers and task predictors, such that they can be fine-tuned efficiently on new datasets or meta-tasks. We demonstrate the efficacy of the proposed task-specific, adaptable IQA approach, using two clinical applications for ultrasound-guided prostate intervention and pneumonia detection on X-ray images.
* Accepted to Medical Image Analysis; Final published version available at: https://doi.org/10.1016/j.media.2022.102427
Image quality assessment by overlapping task-specific and task-agnostic measures: application to prostate multiparametric MR images for cancer segmentation
Feb 20, 2022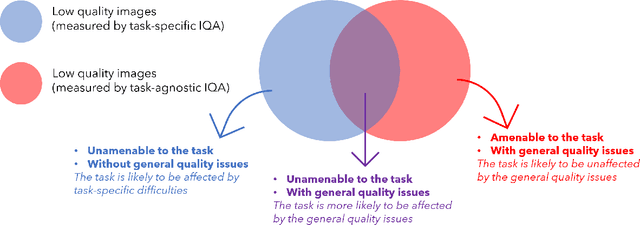
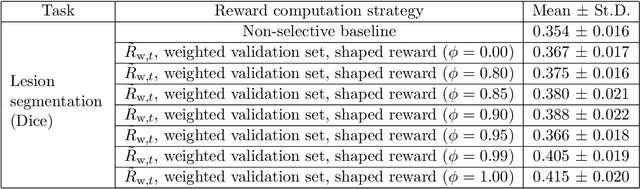

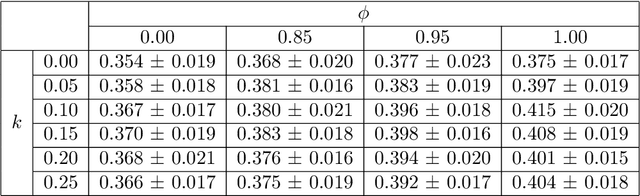
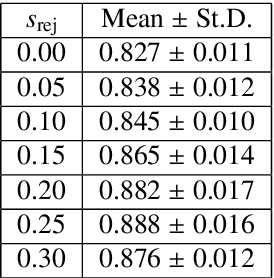
Abstract:Image quality assessment (IQA) in medical imaging can be used to ensure that downstream clinical tasks can be reliably performed. Quantifying the impact of an image on the specific target tasks, also named as task amenability, is needed. A task-specific IQA has recently been proposed to learn an image-amenability-predicting controller simultaneously with a target task predictor. This allows for the trained IQA controller to measure the impact an image has on the target task performance, when this task is performed using the predictor, e.g. segmentation and classification neural networks in modern clinical applications. In this work, we propose an extension to this task-specific IQA approach, by adding a task-agnostic IQA based on auto-encoding as the target task. Analysing the intersection between low-quality images, deemed by both the task-specific and task-agnostic IQA, may help to differentiate the underpinning factors that caused the poor target task performance. For example, common imaging artefacts may not adversely affect the target task, which would lead to a low task-agnostic quality and a high task-specific quality, whilst individual cases considered clinically challenging, which can not be improved by better imaging equipment or protocols, is likely to result in a high task-agnostic quality but a low task-specific quality. We first describe a flexible reward shaping strategy which allows for the adjustment of weighting between task-agnostic and task-specific quality scoring. Furthermore, we evaluate the proposed algorithm using a clinically challenging target task of prostate tumour segmentation on multiparametric magnetic resonance (mpMR) images, from 850 patients. The proposed reward shaping strategy, with appropriately weighted task-specific and task-agnostic qualities, successfully identified samples that need re-acquisition due to defected imaging process.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge