Stephen P. Pereira
Competing for pixels: a self-play algorithm for weakly-supervised segmentation
May 26, 2024Abstract:Weakly-supervised segmentation (WSS) methods, reliant on image-level labels indicating object presence, lack explicit correspondence between labels and regions of interest (ROIs), posing a significant challenge. Despite this, WSS methods have attracted attention due to their much lower annotation costs compared to fully-supervised segmentation. Leveraging reinforcement learning (RL) self-play, we propose a novel WSS method that gamifies image segmentation of a ROI. We formulate segmentation as a competition between two agents that compete to select ROI-containing patches until exhaustion of all such patches. The score at each time-step, used to compute the reward for agent training, represents likelihood of object presence within the selection, determined by an object presence detector pre-trained using only image-level binary classification labels of object presence. Additionally, we propose a game termination condition that can be called by either side upon exhaustion of all ROI-containing patches, followed by the selection of a final patch from each. Upon termination, the agent is incentivised if ROI-containing patches are exhausted or disincentivised if an ROI-containing patch is found by the competitor. This competitive setup ensures minimisation of over- or under-segmentation, a common problem with WSS methods. Extensive experimentation across four datasets demonstrates significant performance improvements over recent state-of-the-art methods. Code: https://github.com/s-sd/spurl/tree/main/wss
Active learning using adaptable task-based prioritisation
Dec 03, 2022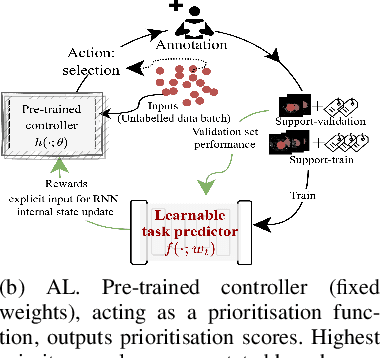
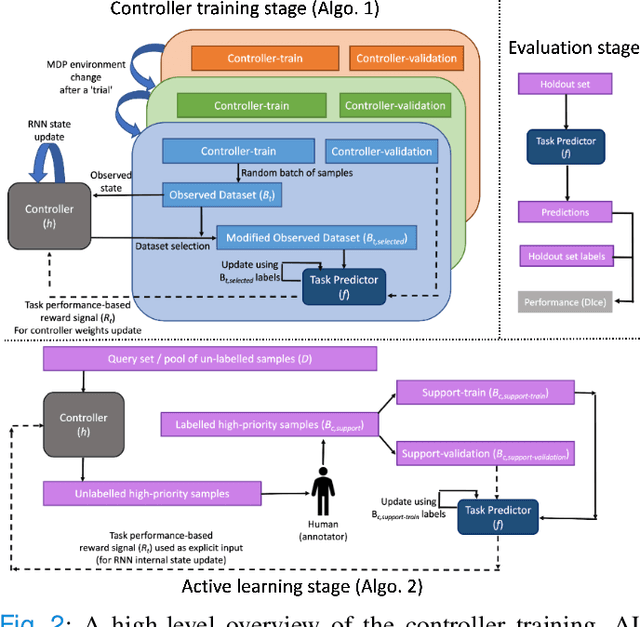
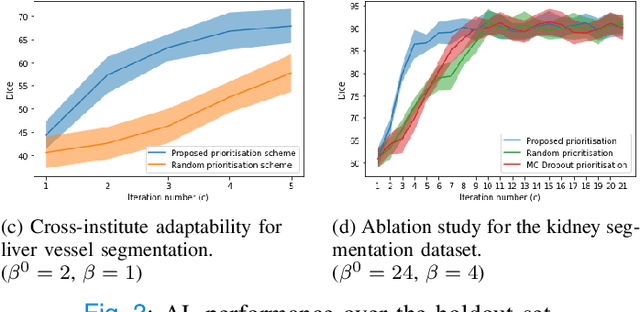
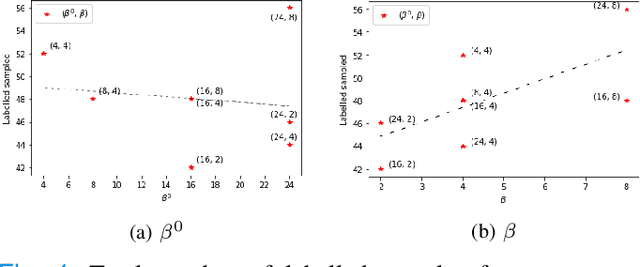
Abstract:Supervised machine learning-based medical image computing applications necessitate expert label curation, while unlabelled image data might be relatively abundant. Active learning methods aim to prioritise a subset of available image data for expert annotation, for label-efficient model training. We develop a controller neural network that measures priority of images in a sequence of batches, as in batch-mode active learning, for multi-class segmentation tasks. The controller is optimised by rewarding positive task-specific performance gain, within a Markov decision process (MDP) environment that also optimises the task predictor. In this work, the task predictor is a segmentation network. A meta-reinforcement learning algorithm is proposed with multiple MDPs, such that the pre-trained controller can be adapted to a new MDP that contains data from different institutes and/or requires segmentation of different organs or structures within the abdomen. We present experimental results using multiple CT datasets from more than one thousand patients, with segmentation tasks of nine different abdominal organs, to demonstrate the efficacy of the learnt prioritisation controller function and its cross-institute and cross-organ adaptability. We show that the proposed adaptable prioritisation metric yields converging segmentation accuracy for the novel class of kidney, unseen in training, using between approximately 40\% to 60\% of labels otherwise required with other heuristic or random prioritisation metrics. For clinical datasets of limited size, the proposed adaptable prioritisation offers a performance improvement of 22.6\% and 10.2\% in Dice score, for tasks of kidney and liver vessel segmentation, respectively, compared to random prioritisation and alternative active sampling strategies.
Voice-assisted Image Labelling for Endoscopic Ultrasound Classification using Neural Networks
Oct 12, 2021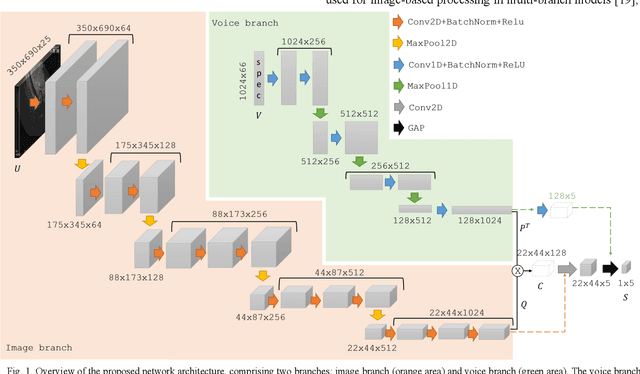
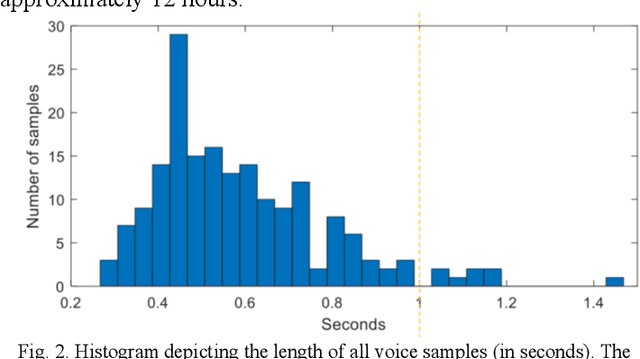
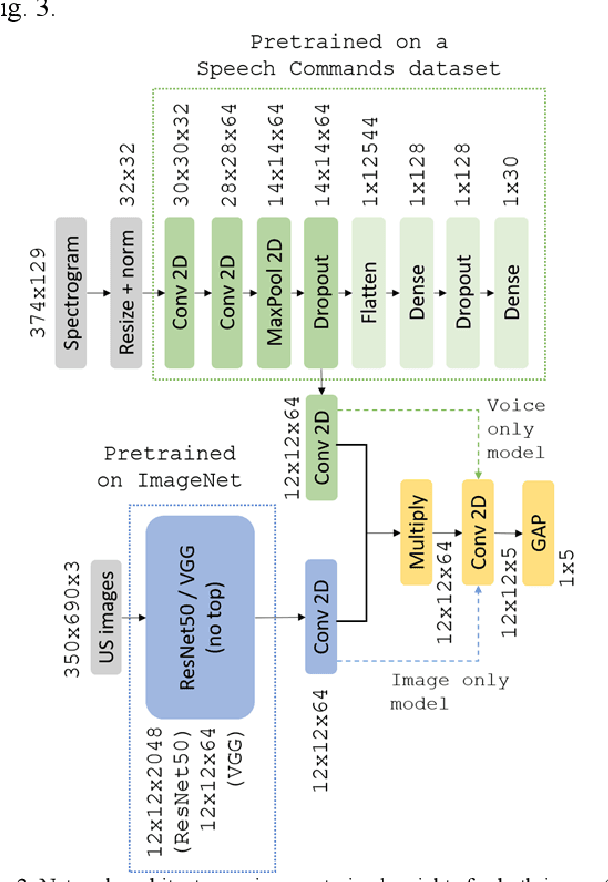
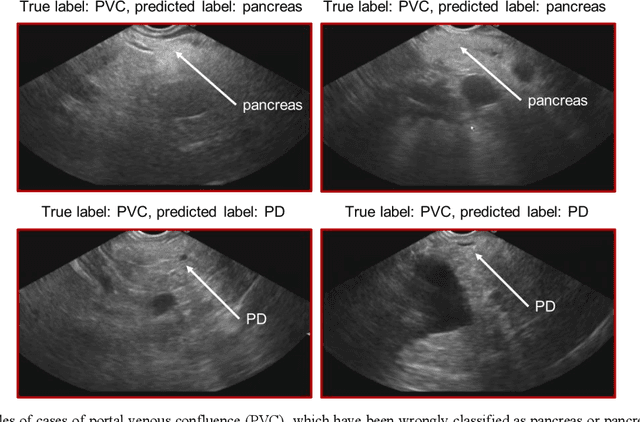
Abstract:Ultrasound imaging is a commonly used technology for visualising patient anatomy in real-time during diagnostic and therapeutic procedures. High operator dependency and low reproducibility make ultrasound imaging and interpretation challenging with a steep learning curve. Automatic image classification using deep learning has the potential to overcome some of these challenges by supporting ultrasound training in novices, as well as aiding ultrasound image interpretation in patient with complex pathology for more experienced practitioners. However, the use of deep learning methods requires a large amount of data in order to provide accurate results. Labelling large ultrasound datasets is a challenging task because labels are retrospectively assigned to 2D images without the 3D spatial context available in vivo or that would be inferred while visually tracking structures between frames during the procedure. In this work, we propose a multi-modal convolutional neural network (CNN) architecture that labels endoscopic ultrasound (EUS) images from raw verbal comments provided by a clinician during the procedure. We use a CNN composed of two branches, one for voice data and another for image data, which are joined to predict image labels from the spoken names of anatomical landmarks. The network was trained using recorded verbal comments from expert operators. Our results show a prediction accuracy of 76% at image level on a dataset with 5 different labels. We conclude that the addition of spoken commentaries can increase the performance of ultrasound image classification, and eliminate the burden of manually labelling large EUS datasets necessary for deep learning applications.
Zero-shot super-resolution with a physically-motivated downsampling kernel for endomicroscopy
Mar 25, 2021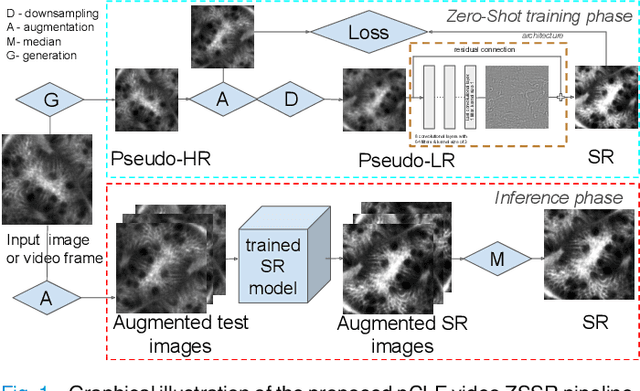
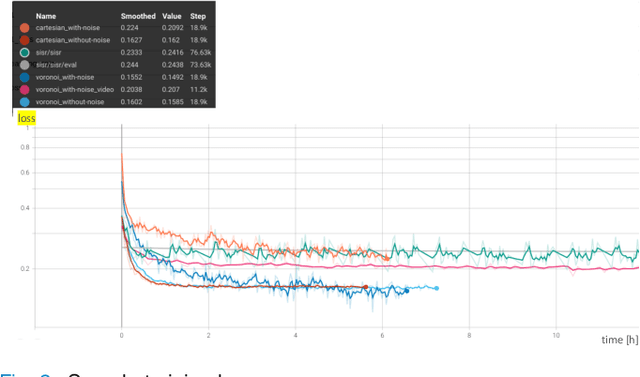
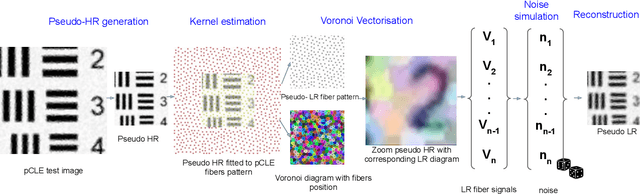
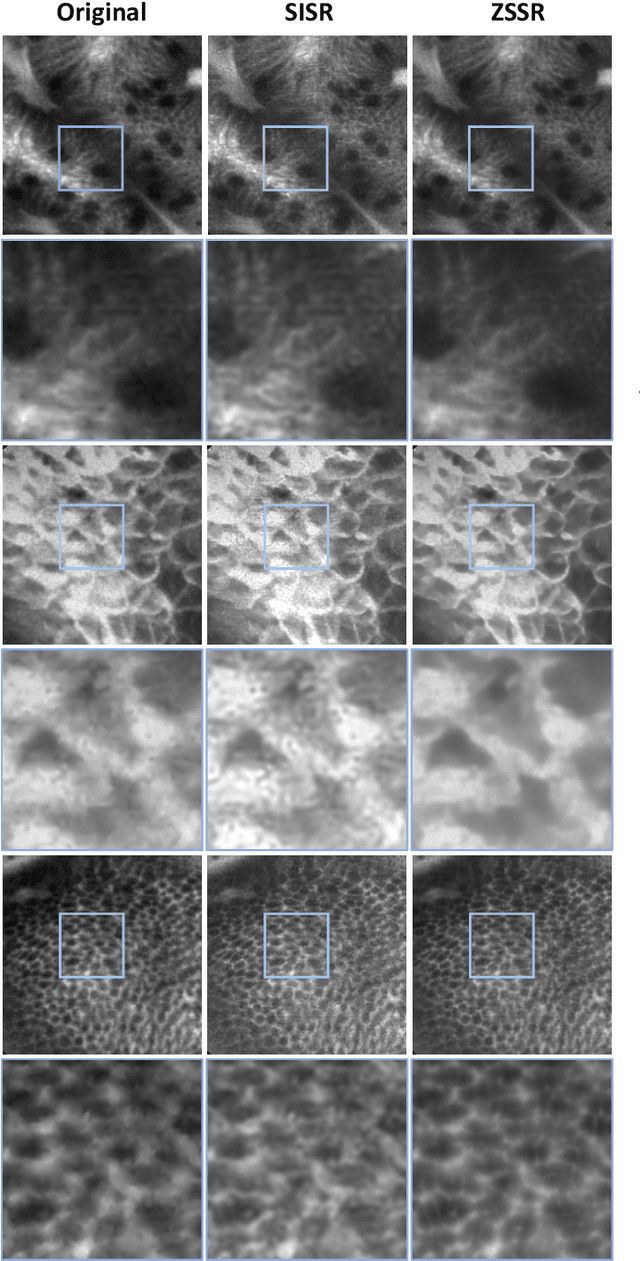
Abstract:Super-resolution (SR) methods have seen significant advances thanks to the development of convolutional neural networks (CNNs). CNNs have been successfully employed to improve the quality of endomicroscopy imaging. Yet, the inherent limitation of research on SR in endomicroscopy remains the lack of ground truth high-resolution (HR) images, commonly used for both supervised training and reference-based image quality assessment (IQA). Therefore, alternative methods, such as unsupervised SR are being explored. To address the need for non-reference image quality improvement, we designed a novel zero-shot super-resolution (ZSSR) approach that relies only on the endomicroscopy data to be processed in a self-supervised manner without the need for ground-truth HR images. We tailored the proposed pipeline to the idiosyncrasies of endomicroscopy by introducing both: a physically-motivated Voronoi downscaling kernel accounting for the endomicroscope's irregular fibre-based sampling pattern, and realistic noise patterns. We also took advantage of video sequences to exploit a sequence of images for self-supervised zero-shot image quality improvement. We run ablation studies to assess our contribution in regards to the downscaling kernel and noise simulation. We validate our methodology on both synthetic and original data. Synthetic experiments were assessed with reference-based IQA, while our results for original images were evaluated in a user study conducted with both expert and non-expert observers. The results demonstrated superior performance in image quality of ZSSR reconstructions in comparison to the baseline method. The ZSSR is also competitive when compared to supervised single-image SR, especially being the preferred reconstruction technique by experts.
Learning from Irregularly Sampled Data for Endomicroscopy Super-resolution: A Comparative Study of Sparse and Dense Approaches
Nov 29, 2019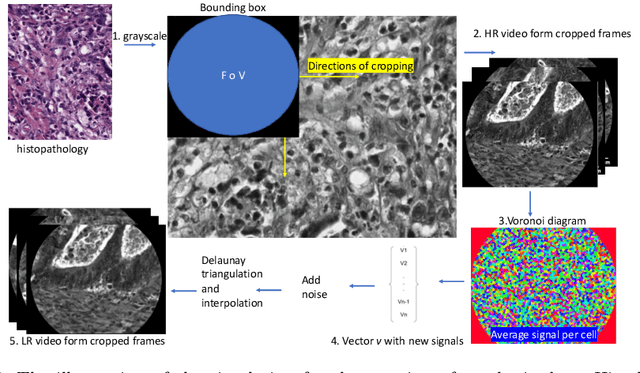
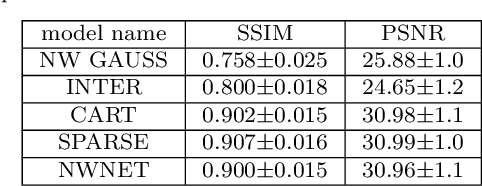
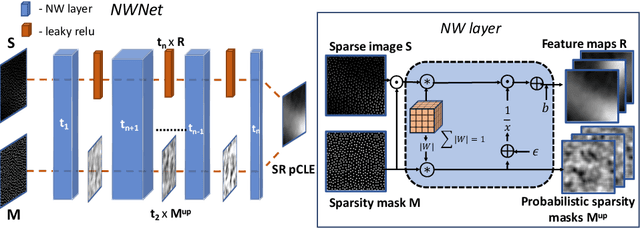
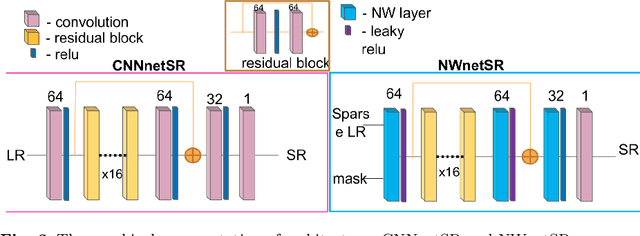
Abstract:Purpose: Probe-based Confocal Laser Endomicroscopy (pCLE) enables performing an optical biopsy, providing real-time microscopic images, via a probe. pCLE probes consist of multiple optical fibres arranged in a bundle, which taken together generate signals in an irregularly sampled pattern. Current pCLE reconstruction is based on interpolating irregular signals onto an over-sampled Cartesian grid, using a naive linear interpolation. It was shown that Convolutional Neural Networks (CNNs) could improve pCLE image quality. Although classical CNNs were applied to pCLE, input data were limited to reconstructed images in contrast to irregular data produced by pCLE. Methods: We compare pCLE reconstruction and super-resolution (SR) methods taking irregularly sampled or reconstructed pCLE images as input. We also propose to embed a Nadaraya-Watson (NW) kernel regression into the CNN framework as a novel trainable CNN layer. Using the NW layer and exemplar-based super-resolution, we design an NWNetSR architecture that allows for reconstructing high-quality pCLE images directly from the irregularly sampled input data. We created synthetic sparse pCLE images to evaluate our methodology. Results: The results were validated through an image quality assessment based on a combination of the following metrics: Peak signal-to-noise ratio, the Structural Similarity Index. Conclusion: Both dense and sparse CNNs outperform the reconstruction method currently used in the clinic. The main contributions of our study are a comparison of sparse and dense approach in pCLE image reconstruction, implementing trainable generalised NW kernel regression, and adaptation of synthetic data for training pCLE SR.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge