Yipei Wang
On the Effects of Adversarial Perturbations on Distribution Robustness
Jan 23, 2026Abstract:Adversarial robustness refers to a model's ability to resist perturbation of inputs, while distribution robustness evaluates the performance of the model under data shifts. Although both aim to ensure reliable performance, prior work has revealed a tradeoff in distribution and adversarial robustness. Specifically, adversarial training might increase reliance on spurious features, which can harm distribution robustness, especially the performance on some underrepresented subgroups. We present a theoretical analysis of adversarial and distribution robustness that provides a tractable surrogate for per-step adversarial training by studying models trained on perturbed data. In addition to the tradeoff, our work further identified a nuanced phenomenon that $\ell_\infty$ perturbations on data with moderate bias can yield an increase in distribution robustness. Moreover, the gain in distribution robustness remains on highly skewed data when simplicity bias induces reliance on the core feature, characterized as greater feature separability. Our theoretical analysis extends the understanding of the tradeoff by highlighting the interplay of the tradeoff and the feature separability. Despite the tradeoff that persists in many cases, overlooking the role of feature separability may lead to misleading conclusions about robustness.
Understanding the Transfer Limits of Vision Foundation Models
Jan 22, 2026Abstract:Foundation models leverage large-scale pretraining to capture extensive knowledge, demonstrating generalization in a wide range of language tasks. By comparison, vision foundation models (VFMs) often exhibit uneven improvements across downstream tasks, despite substantial computational investment. We postulate that this limitation arises from a mismatch between pretraining objectives and the demands of downstream vision-and-imaging tasks. Pretraining strategies like masked image reconstruction or contrastive learning shape representations for tasks such as recovery of generic visual patterns or global semantic structures, which may not align with the task-specific requirements of downstream applications including segmentation, classification, or image synthesis. To investigate this in a concrete real-world clinical area, we assess two VFMs, a reconstruction-focused MAE-based model (ProFound) and a contrastive-learning-based model (ProViCNet), on five prostate multiparametric MR imaging tasks, examining how such task alignment influences transfer performance, i.e., from pretraining to fine-tuning. Our findings indicate that better alignment between pretraining and downstream tasks, measured by simple divergence metrics such as maximum-mean-discrepancy (MMD) between the same features before and after fine-tuning, correlates with greater performance improvements and faster convergence, emphasizing the importance of designing and analyzing pretraining objectives with downstream applicability in mind.
Learning to learn skill assessment for fetal ultrasound scanning
Dec 30, 2025Abstract:Traditionally, ultrasound skill assessment has relied on expert supervision and feedback, a process known for its subjectivity and time-intensive nature. Previous works on quantitative and automated skill assessment have predominantly employed supervised learning methods, often limiting the analysis to predetermined or assumed factors considered influential in determining skill levels. In this work, we propose a novel bi-level optimisation framework that assesses fetal ultrasound skills by how well a task is performed on the acquired fetal ultrasound images, without using manually predefined skill ratings. The framework consists of a clinical task predictor and a skill predictor, which are optimised jointly by refining the two networks simultaneously. We validate the proposed method on real-world clinical ultrasound videos of scanning the fetal head. The results demonstrate the feasibility of predicting ultrasound skills by the proposed framework, which quantifies optimised task performance as a skill indicator.
Policy to Assist Iteratively Local Segmentation: Optimising Modality and Location Selection for Prostate Cancer Localisation
Aug 05, 2025Abstract:Radiologists often mix medical image reading strategies, including inspection of individual modalities and local image regions, using information at different locations from different images independently as well as concurrently. In this paper, we propose a recommend system to assist machine learning-based segmentation models, by suggesting appropriate image portions along with the best modality, such that prostate cancer segmentation performance can be maximised. Our approach trains a policy network that assists tumor localisation, by recommending both the optimal imaging modality and the specific sections of interest for review. During training, a pre-trained segmentation network mimics radiologist inspection on individual or variable combinations of these imaging modalities and their sections - selected by the policy network. Taking the locally segmented regions as an input for the next step, this dynamic decision making process iterates until all cancers are best localised. We validate our method using a data set of 1325 labelled multiparametric MRI images from prostate cancer patients, demonstrating its potential to improve annotation efficiency and segmentation accuracy, especially when challenging pathology is present. Experimental results show that our approach can surpass standard segmentation networks. Perhaps more interestingly, our trained agent independently developed its own optimal strategy, which may or may not be consistent with current radiologist guidelines such as PI-RADS. This observation also suggests a promising interactive application, in which the proposed policy networks assist human radiologists.
Promptable cancer segmentation using minimal expert-curated data
May 23, 2025Abstract:Automated segmentation of cancer on medical images can aid targeted diagnostic and therapeutic procedures. However, its adoption is limited by the high cost of expert annotations required for training and inter-observer variability in datasets. While weakly-supervised methods mitigate some challenges, using binary histology labels for training as opposed to requiring full segmentation, they require large paired datasets of histology and images, which are difficult to curate. Similarly, promptable segmentation aims to allow segmentation with no re-training for new tasks at inference, however, existing models perform poorly on pathological regions, again necessitating large datasets for training. In this work we propose a novel approach for promptable segmentation requiring only 24 fully-segmented images, supplemented by 8 weakly-labelled images, for training. Curating this minimal data to a high standard is relatively feasible and thus issues with the cost and variability of obtaining labels can be mitigated. By leveraging two classifiers, one weakly-supervised and one fully-supervised, our method refines segmentation through a guided search process initiated by a single-point prompt. Our approach outperforms existing promptable segmentation methods, and performs comparably with fully-supervised methods, for the task of prostate cancer segmentation, while using substantially less annotated data (up to 100X less). This enables promptable segmentation with very minimal labelled data, such that the labels can be curated to a very high standard.
Tell2Reg: Establishing spatial correspondence between images by the same language prompts
Feb 05, 2025Abstract:Spatial correspondence can be represented by pairs of segmented regions, such that the image registration networks aim to segment corresponding regions rather than predicting displacement fields or transformation parameters. In this work, we show that such a corresponding region pair can be predicted by the same language prompt on two different images using the pre-trained large multimodal models based on GroundingDINO and SAM. This enables a fully automated and training-free registration algorithm, potentially generalisable to a wide range of image registration tasks. In this paper, we present experimental results using one of the challenging tasks, registering inter-subject prostate MR images, which involves both highly variable intensity and morphology between patients. Tell2Reg is training-free, eliminating the need for costly and time-consuming data curation and labelling that was previously required for this registration task. This approach outperforms unsupervised learning-based registration methods tested, and has a performance comparable to weakly-supervised methods. Additional qualitative results are also presented to suggest that, for the first time, there is a potential correlation between language semantics and spatial correspondence, including the spatial invariance in language-prompted regions and the difference in language prompts between the obtained local and global correspondences. Code is available at https://github.com/yanwenCi/Tell2Reg.git.
T2-Only Prostate Cancer Prediction by Meta-Learning from Bi-Parametric MR Imaging
Nov 11, 2024


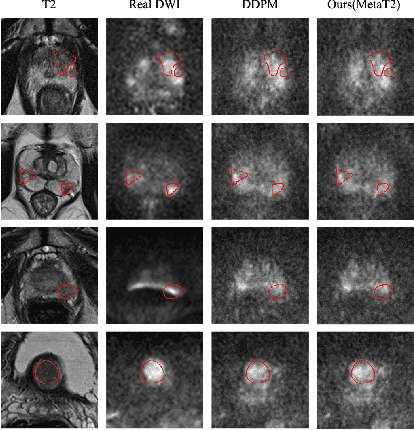
Abstract:Current imaging-based prostate cancer diagnosis requires both MR T2-weighted (T2w) and diffusion-weighted imaging (DWI) sequences, with additional sequences for potentially greater accuracy improvement. However, measuring diffusion patterns in DWI sequences can be time-consuming, prone to artifacts and sensitive to imaging parameters. While machine learning (ML) models have demonstrated radiologist-level accuracy in detecting prostate cancer from these two sequences, this study investigates the potential of ML-enabled methods using only the T2w sequence as input during inference time. We first discuss the technical feasibility of such a T2-only approach, and then propose a novel ML formulation, where DWI sequences - readily available for training purposes - are only used to train a meta-learning model, which subsequently only uses T2w sequences at inference. Using multiple datasets from more than 3,000 prostate cancer patients, we report superior or comparable performance in localising radiologist-identified prostate cancer using our proposed T2-only models, compared with alternative models using T2-only or both sequences as input. Real patient cases are presented and discussed to demonstrate, for the first time, the exclusively true-positive cases from models with different input sequences.
AI-assisted prostate cancer detection and localisation on biparametric MR by classifying radiologist-positives
Oct 30, 2024



Abstract:Prostate cancer diagnosis through MR imaging have currently relied on radiologists' interpretation, whilst modern AI-based methods have been developed to detect clinically significant cancers independent of radiologists. In this study, we propose to develop deep learning models that improve the overall cancer diagnostic accuracy, by classifying radiologist-identified patients or lesions (i.e. radiologist-positives), as opposed to the existing models that are trained to discriminate over all patients. We develop a single voxel-level classification model, with a simple percentage threshold to determine positive cases, at levels of lesions, Barzell-zones and patients. Based on the presented experiments from two clinical data sets, consisting of histopathology-labelled MR images from more than 800 and 500 patients in the respective UCLA and UCL PROMIS studies, we show that the proposed strategy can improve the diagnostic accuracy, by augmenting the radiologist reading of the MR imaging. Among varying definition of clinical significance, the proposed strategy, for example, achieved a specificity of 44.1% (with AI assistance) from 36.3% (by radiologists alone), at a controlled sensitivity of 80.0% on the publicly available UCLA data set. This provides measurable clinical values in a range of applications such as reducing unnecessary biopsies, lowering cost in cancer screening and quantifying risk in therapies.
Students Rather Than Experts: A New AI For Education Pipeline To Model More Human-Like And Personalised Early Adolescences
Oct 21, 2024



Abstract:The capabilities of large language models (LLMs) have been applied in expert systems across various domains, providing new opportunities for AI in Education. Educational interactions involve a cyclical exchange between teachers and students. Current research predominantly focuses on using LLMs to simulate teachers, leveraging their expertise to enhance student learning outcomes. However, the simulation of students, which could improve teachers' instructional skills, has received insufficient attention due to the challenges of modeling and evaluating virtual students. This research asks: Can LLMs be utilized to develop virtual student agents that mimic human-like behavior and individual variability? Unlike expert systems focusing on knowledge delivery, virtual students must replicate learning difficulties, emotional responses, and linguistic uncertainties. These traits present significant challenges in both modeling and evaluation. To address these issues, this study focuses on language learning as a context for modeling virtual student agents. We propose a novel AI4Education framework, called SOE (Scene-Object-Evaluation), to systematically construct LVSA (LLM-based Virtual Student Agents). By curating a dataset of personalized teacher-student interactions with various personality traits, question types, and learning stages, and fine-tuning LLMs using LoRA, we conduct multi-dimensional evaluation experiments. Specifically, we: (1) develop a theoretical framework for generating LVSA; (2) integrate human subjective evaluation metrics into GPT-4 assessments, demonstrating a strong correlation between human evaluators and GPT-4 in judging LVSA authenticity; and (3) validate that LLMs can generate human-like, personalized virtual student agents in educational contexts, laying a foundation for future applications in pre-service teacher training and multi-agent simulation environments.
Can LVLMs Describe Videos like Humans? A Five-in-One Video Annotations Benchmark for Better Human-Machine Comparison
Oct 20, 2024



Abstract:Large vision-language models (LVLMs) have made significant strides in addressing complex video tasks, sparking researchers' interest in their human-like multimodal understanding capabilities. Video description serves as a fundamental task for evaluating video comprehension, necessitating a deep understanding of spatial and temporal dynamics, which presents challenges for both humans and machines. Thus, investigating whether LVLMs can describe videos as comprehensively as humans (through reasonable human-machine comparisons using video captioning as a proxy task) will enhance our understanding and application of these models. However, current benchmarks for video comprehension have notable limitations, including short video durations, brief annotations, and reliance on a single annotator's perspective. These factors hinder a comprehensive assessment of LVLMs' ability to understand complex, lengthy videos and prevent the establishment of a robust human baseline that accurately reflects human video comprehension capabilities. To address these issues, we propose a novel benchmark, FIOVA (Five In One Video Annotations), designed to evaluate the differences between LVLMs and human understanding more comprehensively. FIOVA includes 3,002 long video sequences (averaging 33.6 seconds) that cover diverse scenarios with complex spatiotemporal relationships. Each video is annotated by five distinct annotators, capturing a wide range of perspectives and resulting in captions that are 4-15 times longer than existing benchmarks, thereby establishing a robust baseline that represents human understanding comprehensively for the first time in video description tasks. Using the FIOVA benchmark, we conducted an in-depth evaluation of six state-of-the-art LVLMs, comparing their performance with humans. More detailed information can be found at https://huuuuusy.github.io/fiova/.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge