Yinsong Xu
Locating and Mitigating Gradient Conflicts in Point Cloud Domain Adaptation via Saliency Map Skewness
Apr 22, 2025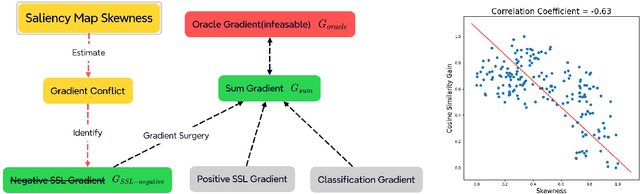
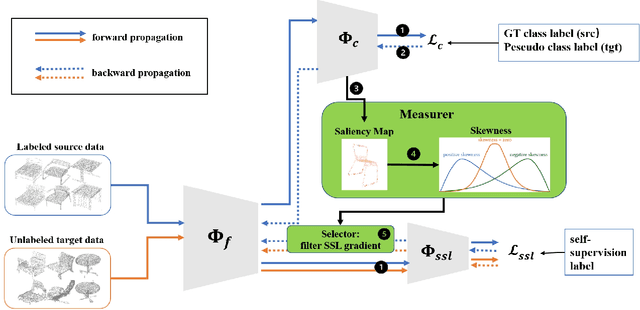
Abstract:Object classification models utilizing point cloud data are fundamental for 3D media understanding, yet they often struggle with unseen or out-of-distribution (OOD) scenarios. Existing point cloud unsupervised domain adaptation (UDA) methods typically employ a multi-task learning (MTL) framework that combines primary classification tasks with auxiliary self-supervision tasks to bridge the gap between cross-domain feature distributions. However, our further experiments demonstrate that not all gradients from self-supervision tasks are beneficial and some may negatively impact the classification performance. In this paper, we propose a novel solution, termed Saliency Map-based Data Sampling Block (SM-DSB), to mitigate these gradient conflicts. Specifically, our method designs a new scoring mechanism based on the skewness of 3D saliency maps to estimate gradient conflicts without requiring target labels. Leveraging this, we develop a sample selection strategy that dynamically filters out samples whose self-supervision gradients are not beneficial for the classification. Our approach is scalable, introducing modest computational overhead, and can be integrated into all the point cloud UDA MTL frameworks. Extensive evaluations demonstrate that our method outperforms state-of-the-art approaches. In addition, we provide a new perspective on understanding the UDA problem through back-propagation analysis.
Poisson Ordinal Network for Gleason Group Estimation Using Bi-Parametric MRI
Jul 08, 2024



Abstract:The Gleason groups serve as the primary histological grading system for prostate cancer, providing crucial insights into the cancer's potential for growth and metastasis. In clinical practice, pathologists determine the Gleason groups based on specimens obtained from ultrasound-guided biopsies. In this study, we investigate the feasibility of directly estimating the Gleason groups from MRI scans to reduce otherwise required biopsies. We identify two characteristics of this task, ordinality and the resulting dependent yet unknown variances between Gleason groups. In addition to the inter- / intra- observer variability in a multi-step Gleason scoring process based on the interpretation of Gleason patterns, our MR-based prediction is also subject to specimen sampling variance and, to a lesser degree, varying MR imaging protocols. To address this challenge, we propose a novel Poisson ordinal network (PON). PONs model the prediction using a Poisson distribution and leverages Poisson encoding and Poisson focal loss to capture a learnable dependency between ordinal classes (here, Gleason groups), rather than relying solely on the numerical ground-truth (e.g. Gleason Groups 1-5 or Gleason Scores 6-10). To improve this modelling efficacy, PONs also employ contrastive learning with a memory bank to regularise intra-class variance, decoupling the memory requirement of contrast learning from the batch size. Experimental results based on the images labelled by saturation biopsies from 265 prior-biopsy-blind patients, across two tasks demonstrate the superiority and effectiveness of our proposed method.
EviPrompt: A Training-Free Evidential Prompt Generation Method for Segment Anything Model in Medical Images
Nov 10, 2023



Abstract:Medical image segmentation has immense clinical applicability but remains a challenge despite advancements in deep learning. The Segment Anything Model (SAM) exhibits potential in this field, yet the requirement for expertise intervention and the domain gap between natural and medical images poses significant obstacles. This paper introduces a novel training-free evidential prompt generation method named EviPrompt to overcome these issues. The proposed method, built on the inherent similarities within medical images, requires only a single reference image-annotation pair, making it a training-free solution that significantly reduces the need for extensive labeling and computational resources. First, to automatically generate prompts for SAM in medical images, we introduce an evidential method based on uncertainty estimation without the interaction of clinical experts. Then, we incorporate the human prior into the prompts, which is vital for alleviating the domain gap between natural and medical images and enhancing the applicability and usefulness of SAM in medical scenarios. EviPrompt represents an efficient and robust approach to medical image segmentation, with evaluations across a broad range of tasks and modalities confirming its efficacy.
Incorporating Pre-training Data Matters in Unsupervised Domain Adaptation
Aug 06, 2023Abstract:Unsupervised domain adaptation(UDA) and Source-free UDA(SFUDA) methods formulate the problem involving two domains: source and target. They typically employ a standard training approach that begins with models pre-trained on large-scale datasets e.g., ImageNet, while rarely discussing its effect. Recognizing this gap, we investigate the following research questions: (1) What is the correlation among ImageNet, the source, and the target domain? (2) How does pre-training on ImageNet influence the target risk? To answer the first question, we empirically observed an interesting Spontaneous Pulling (SP) Effect in fine-tuning where the discrepancies between any two of the three domains (ImageNet, Source, Target) decrease but at the cost of the impaired semantic structure of the pre-train domain. For the second question, we put forward a theory to explain SP and quantify that the target risk is bound by gradient disparities among the three domains. Our observations reveal a key limitation of existing methods: it hinders the adaptation performance if the semantic cluster structure of the pre-train dataset (i.e.ImageNet) is impaired. To address it, we incorporate ImageNet as the third domain and redefine the UDA/SFUDA as a three-player game. Specifically, inspired by the theory and empirical findings, we present a novel framework termed TriDA which additionally preserves the semantic structure of the pre-train dataset during fine-tuning. Experimental results demonstrate that it achieves state-of-the-art performance across various UDA and SFUDA benchmarks.
Delving into the Continuous Domain Adaptation
Aug 28, 2022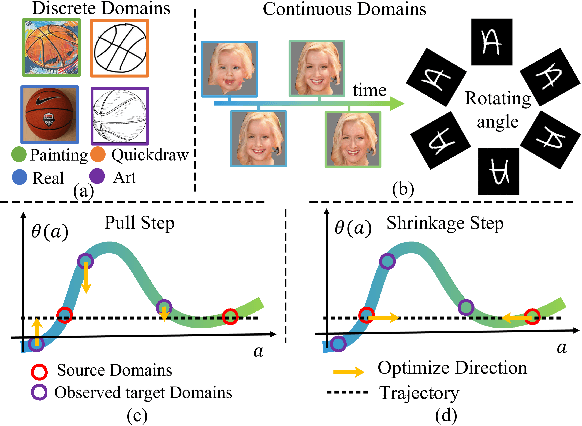
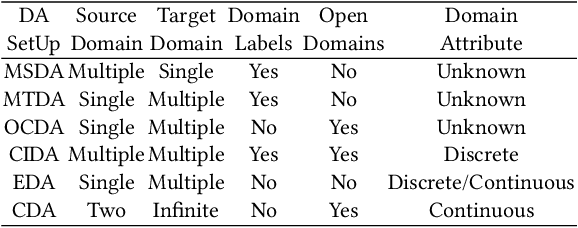
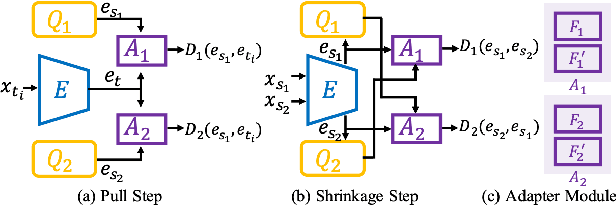
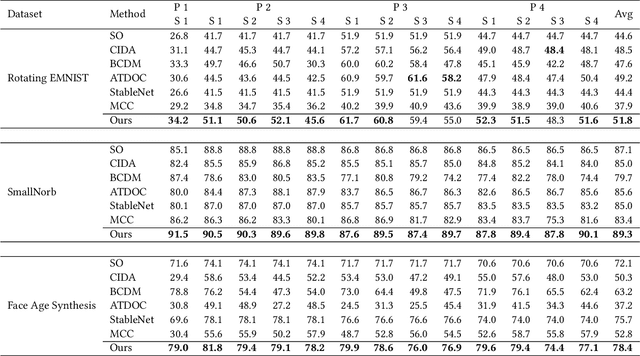
Abstract:Existing domain adaptation methods assume that domain discrepancies are caused by a few discrete attributes and variations, e.g., art, real, painting, quickdraw, etc. We argue that this is not realistic as it is implausible to define the real-world datasets using a few discrete attributes. Therefore, we propose to investigate a new problem namely the Continuous Domain Adaptation (CDA) through the lens where infinite domains are formed by continuously varying attributes. Leveraging knowledge of two labeled source domains and several observed unlabeled target domains data, the objective of CDA is to learn a generalized model for whole data distribution with the continuous attribute. Besides the contributions of formulating a new problem, we also propose a novel approach as a strong CDA baseline. To be specific, firstly we propose a novel alternating training strategy to reduce discrepancies among multiple domains meanwhile generalize to unseen target domains. Secondly, we propose a continuity constraint when estimating the cross-domain divergence measurement. Finally, to decouple the discrepancy from the mini-batch size, we design a domain-specific queue to maintain the global view of the source domain that further boosts the adaptation performances. Our method is proven to achieve the state-of-the-art in CDA problem using extensive experiments. The code is available at https://github.com/SPIresearch/CDA.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge