Yiwen Li
CyberBOT: Towards Reliable Cybersecurity Education via Ontology-Grounded Retrieval Augmented Generation
Apr 01, 2025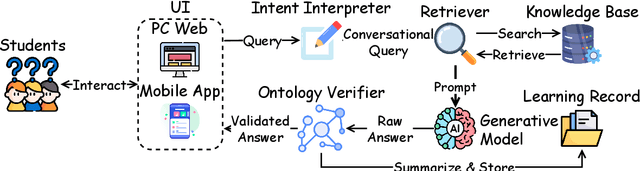
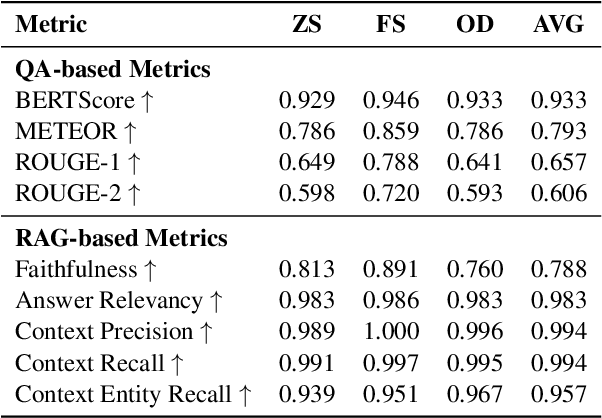

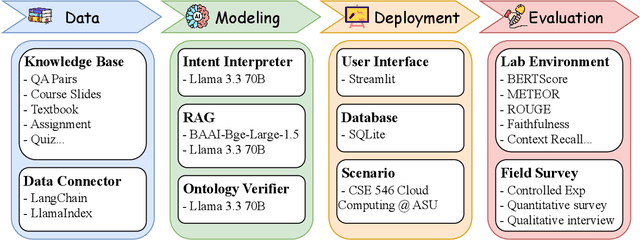
Abstract:Advancements in large language models (LLMs) have enabled the development of intelligent educational tools that support inquiry-based learning across technical domains. In cybersecurity education, where accuracy and safety are paramount, systems must go beyond surface-level relevance to provide information that is both trustworthy and domain-appropriate. To address this challenge, we introduce CyberBOT, a question-answering chatbot that leverages a retrieval-augmented generation (RAG) pipeline to incorporate contextual information from course-specific materials and validate responses using a domain-specific cybersecurity ontology. The ontology serves as a structured reasoning layer that constrains and verifies LLM-generated answers, reducing the risk of misleading or unsafe guidance. CyberBOT has been deployed in a large graduate-level course at Arizona State University (ASU), where more than one hundred students actively engage with the system through a dedicated web-based platform. Computational evaluations in lab environments highlight the potential capacity of CyberBOT, and a forthcoming field study will evaluate its pedagogical impact. By integrating structured domain reasoning with modern generative capabilities, CyberBOT illustrates a promising direction for developing reliable and curriculum-aligned AI applications in specialized educational contexts.
'One size doesn't fit all': Learning how many Examples to use for In-Context Learning for Improved Text Classification
Mar 11, 2024Abstract:Predictive models in natural language processing (NLP) have evolved from training models from scratch to fine-tuning pre-trained models with labelled data. An extreme form of this fine-tuning involves in-context learning (ICL), where the output of a pre-trained generative model (frozen decoder parameters) is controlled only with variations in the input strings (called instructions or prompts). An important component of ICL is the use of a small number of labelled data instances as examples in the prompt. While existing work uses a static number of examples during inference for each data instance, in this paper we propose a novel methodology of dynamically adapting the number of examples as per the data. This is analogous to the use of a variable-sized neighborhood in k-nearest neighbors (k-NN) classifier. In our proposed workflow of adaptive ICL (AICL), the number of demonstrations to employ during the inference on a particular data instance is predicted by the Softmax posteriors of a classifier. The parameters of this classifier are fitted on the optimal number of examples in ICL required to correctly infer the label of each instance in the training set with the hypothesis that a test instance that is similar to a training instance should use the same (or a closely matching) number of few-shot examples. Our experiments show that our AICL method results in improvement in text classification task on several standard datasets.
Semi-weakly-supervised neural network training for medical image registration
Feb 16, 2024



Abstract:For training registration networks, weak supervision from segmented corresponding regions-of-interest (ROIs) have been proven effective for (a) supplementing unsupervised methods, and (b) being used independently in registration tasks in which unsupervised losses are unavailable or ineffective. This correspondence-informing supervision entails cost in annotation that requires significant specialised effort. This paper describes a semi-weakly-supervised registration pipeline that improves the model performance, when only a small corresponding-ROI-labelled dataset is available, by exploiting unlabelled image pairs. We examine two types of augmentation methods by perturbation on network weights and image resampling, such that consistency-based unsupervised losses can be applied on unlabelled data. The novel WarpDDF and RegCut approaches are proposed to allow commutative perturbation between an image pair and the predicted spatial transformation (i.e. respective input and output of registration networks), distinct from existing perturbation methods for classification or segmentation. Experiments using 589 male pelvic MR images, labelled with eight anatomical ROIs, show the improvement in registration performance and the ablated contributions from the individual strategies. Furthermore, this study attempts to construct one of the first computational atlases for pelvic structures, enabled by registering inter-subject MRs, and quantifies the significant differences due to the proposed semi-weak supervision with a discussion on the potential clinical use of example atlas-derived statistics.
A Recycling Training Strategy for Medical Image Segmentation with Diffusion Denoising Models
Aug 30, 2023



Abstract:Denoising diffusion models have found applications in image segmentation by generating segmented masks conditioned on images. Existing studies predominantly focus on adjusting model architecture or improving inference such as test-time sampling strategies. In this work, we focus on training strategy improvements and propose a novel recycling method. During each training step, a segmentation mask is first predicted given an image and a random noise. This predicted mask, replacing the conventional ground truth mask, is used for denoising task during training. This approach can be interpreted as aligning the training strategy with inference by eliminating the dependence on ground truth masks for generating noisy samples. Our proposed method significantly outperforms standard diffusion training, self-conditioning, and existing recycling strategies across multiple medical imaging data sets: muscle ultrasound, abdominal CT, prostate MR, and brain MR. This holds true for two widely adopted sampling strategies: denoising diffusion probabilistic model and denoising diffusion implicit model. Importantly, existing diffusion models often display a declining or unstable performance during inference, whereas our novel recycling consistently enhances or maintains performance. Furthermore, we show for the first time that, under a fair comparison with the same network architectures and computing budget, the proposed recycling-based diffusion models achieved on-par performance with non-diffusion-based supervised training. This paper summarises these quantitative results and discusses their values, with a fully reproducible JAX-based implementation, released at https://github.com/mathpluscode/ImgX-DiffSeg.
Importance of Aligning Training Strategy with Evaluation for Diffusion Models in 3D Multiclass Segmentation
Mar 10, 2023Abstract:Recently, denoising diffusion probabilistic models (DDPM) have been applied to image segmentation by generating segmentation masks conditioned on images, while the applications were mainly limited to 2D networks without exploiting potential benefits from the 3D formulation. In this work, for the first time, DDPMs are used for 3D multiclass image segmentation. We make three key contributions that all focus on aligning the training strategy with the evaluation methodology, and improving efficiency. Firstly, the model predicts segmentation masks instead of sampled noise and is optimised directly via Dice loss. Secondly, the predicted mask in the previous time step is recycled to generate noise-corrupted masks to reduce information leakage. Finally, the diffusion process during training was reduced to five steps, the same as the evaluation. Through studies on two large multiclass data sets (prostate MR and abdominal CT), we demonstrated significantly improved performance compared to existing DDPMs, and reached competitive performance with non-diffusion segmentation models, based on U-net, within the same compute budget. The JAX-based diffusion framework has been released on https://github.com/mathpluscode/ImgX-DiffSeg.
MONAI: An open-source framework for deep learning in healthcare
Nov 04, 2022



Abstract:Artificial Intelligence (AI) is having a tremendous impact across most areas of science. Applications of AI in healthcare have the potential to improve our ability to detect, diagnose, prognose, and intervene on human disease. For AI models to be used clinically, they need to be made safe, reproducible and robust, and the underlying software framework must be aware of the particularities (e.g. geometry, physiology, physics) of medical data being processed. This work introduces MONAI, a freely available, community-supported, and consortium-led PyTorch-based framework for deep learning in healthcare. MONAI extends PyTorch to support medical data, with a particular focus on imaging, and provide purpose-specific AI model architectures, transformations and utilities that streamline the development and deployment of medical AI models. MONAI follows best practices for software-development, providing an easy-to-use, robust, well-documented, and well-tested software framework. MONAI preserves the simple, additive, and compositional approach of its underlying PyTorch libraries. MONAI is being used by and receiving contributions from research, clinical and industrial teams from around the world, who are pursuing applications spanning nearly every aspect of healthcare.
Prototypical few-shot segmentation for cross-institution male pelvic structures with spatial registration
Sep 13, 2022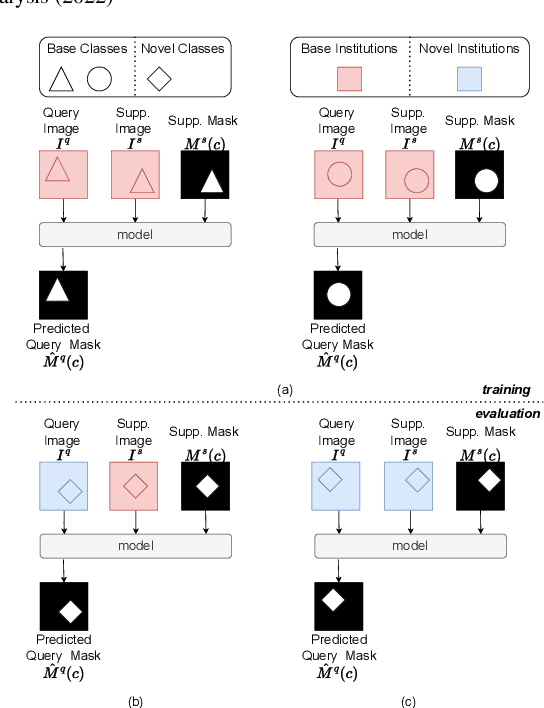

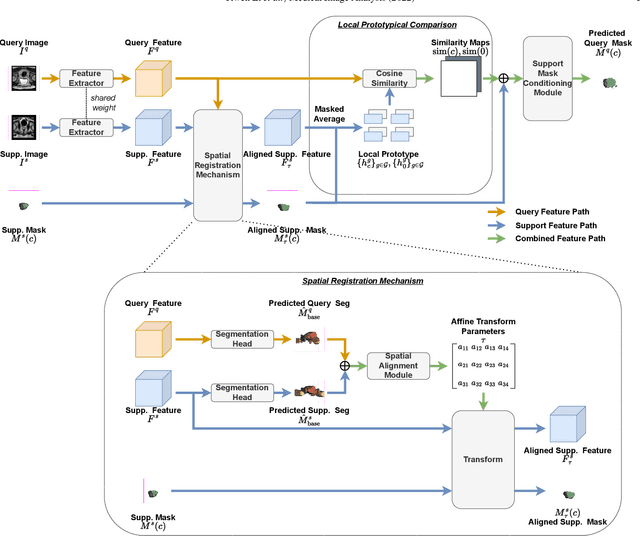
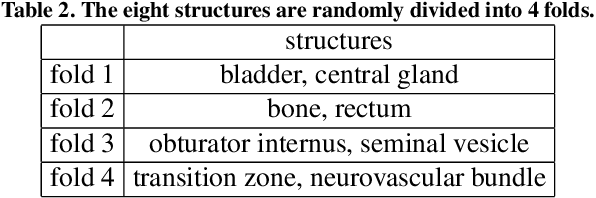
Abstract:The prowess that makes few-shot learning desirable in medical image analysis is the efficient use of the support image data, which are labelled to classify or segment new classes, a task that otherwise requires substantially more training images and expert annotations. This work describes a fully 3D prototypical few-shot segmentation algorithm, such that the trained networks can be effectively adapted to clinically interesting structures that are absent in training, using only a few labelled images from a different institute. First, to compensate for the widely recognised spatial variability between institutions in episodic adaptation of novel classes, a novel spatial registration mechanism is integrated into prototypical learning, consisting of a segmentation head and an spatial alignment module. Second, to assist the training with observed imperfect alignment, support mask conditioning module is proposed to further utilise the annotation available from the support images. Extensive experiments are presented in an application of segmenting eight anatomical structures important for interventional planning, using a data set of 589 pelvic T2-weighted MR images, acquired at seven institutes. The results demonstrate the efficacy in each of the 3D formulation, the spatial registration, and the support mask conditioning, all of which made positive contributions independently or collectively. Compared with the previously proposed 2D alternatives, the few-shot segmentation performance was improved with statistical significance, regardless whether the support data come from the same or different institutes.
Few-shot image segmentation for cross-institution male pelvic organs using registration-assisted prototypical learning
Jan 17, 2022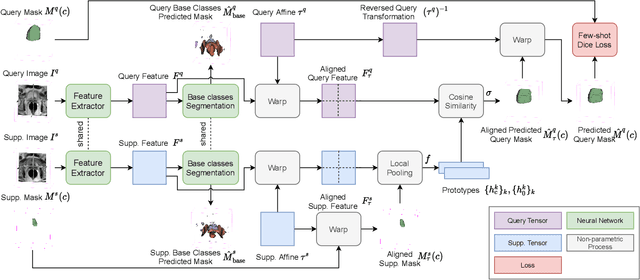
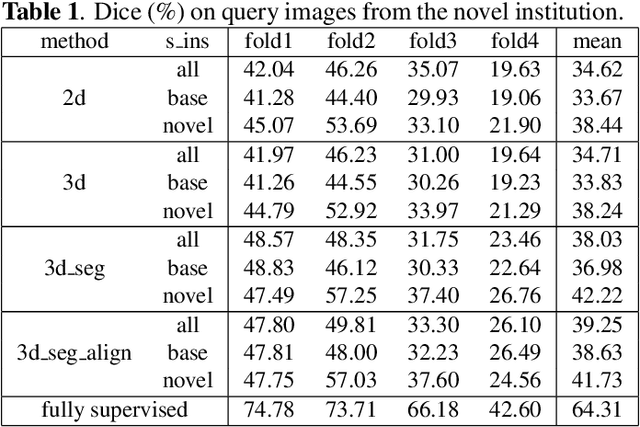
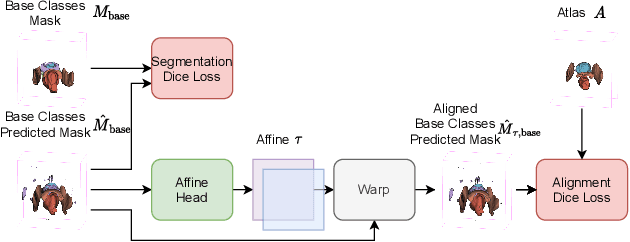
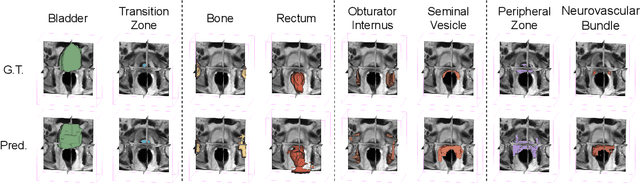
Abstract:The ability to adapt medical image segmentation networks for a novel class such as an unseen anatomical or pathological structure, when only a few labelled examples of this class are available from local healthcare providers, is sought-after. This potentially addresses two widely recognised limitations in deploying modern deep learning models to clinical practice, expertise-and-labour-intensive labelling and cross-institution generalisation. This work presents the first 3D few-shot interclass segmentation network for medical images, using a labelled multi-institution dataset from prostate cancer patients with eight regions of interest. We propose an image alignment module registering the predicted segmentation of both query and support data, in a standard prototypical learning algorithm, to a reference atlas space. The built-in registration mechanism can effectively utilise the prior knowledge of consistent anatomy between subjects, regardless whether they are from the same institution or not. Experimental results demonstrated that the proposed registration-assisted prototypical learning significantly improved segmentation accuracy (p-values<0.01) on query data from a holdout institution, with varying availability of support data from multiple institutions. We also report the additional benefits of the proposed 3D networks with 75% fewer parameters and an arguably simpler implementation, compared with existing 2D few-shot approaches that segment 2D slices of volumetric medical images.
Few-shot Semantic Segmentation with Self-supervision from Pseudo-classes
Oct 22, 2021



Abstract:Despite the success of deep learning methods for semantic segmentation, few-shot semantic segmentation remains a challenging task due to the limited training data and the generalisation requirement for unseen classes. While recent progress has been particularly encouraging, we discover that existing methods tend to have poor performance in terms of meanIoU when query images contain other semantic classes besides the target class. To address this issue, we propose a novel self-supervised task that generates random pseudo-classes in the background of the query images, providing extra training data that would otherwise be unavailable when predicting individual target classes. To that end, we adopted superpixel segmentation for generating the pseudo-classes. With this extra supervision, we improved the meanIoU performance of the state-of-the-art method by 2.5% and 5.1% on the one-shot tasks, as well as 6.7% and 4.4% on the five-shot tasks, on the PASCAL-5i and COCO benchmarks, respectively.
GroSS: Group-Size Series Decomposition for Whole Search-Space Training
Dec 02, 2019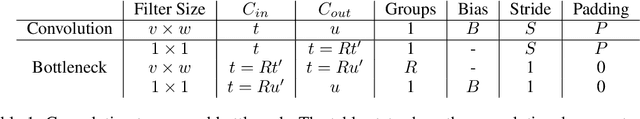
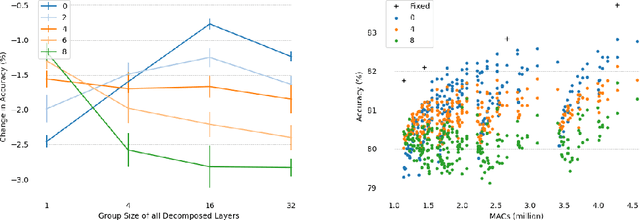
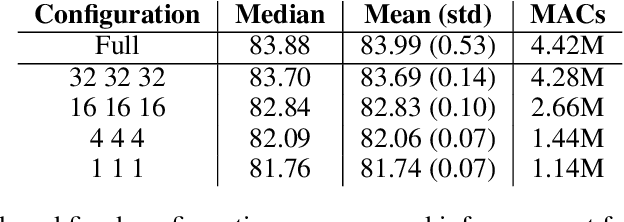
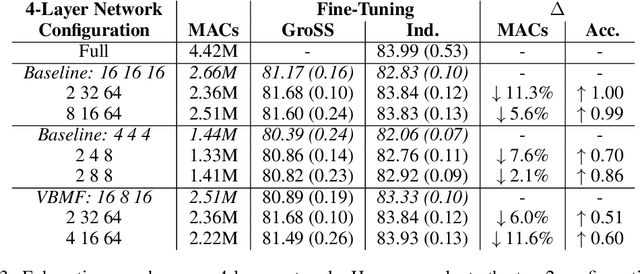
Abstract:We present Group-size Series (GroSS) decomposition, a mathematical formulation of tensor factorisation into a series of approximations of increasing rank terms. GroSS allows for dynamic and differentiable selection of factorisation rank, which is analogous to a grouped convolution. Therefore, to the best of our knowledge, GroSS is the first method to simultaneously train differing numbers of groups within a single layer, as well as all possible combinations between layers. In doing so, GroSS trains an entire grouped convolution architecture search-space concurrently. We demonstrate this through proof-of-concept architecture searches with performance objectives. GroSS represents a significant step towards liberating network architecture search from the burden of training and fine-tuning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge