Qianye Yang
Learning to learn skill assessment for fetal ultrasound scanning
Dec 30, 2025Abstract:Traditionally, ultrasound skill assessment has relied on expert supervision and feedback, a process known for its subjectivity and time-intensive nature. Previous works on quantitative and automated skill assessment have predominantly employed supervised learning methods, often limiting the analysis to predetermined or assumed factors considered influential in determining skill levels. In this work, we propose a novel bi-level optimisation framework that assesses fetal ultrasound skills by how well a task is performed on the acquired fetal ultrasound images, without using manually predefined skill ratings. The framework consists of a clinical task predictor and a skill predictor, which are optimised jointly by refining the two networks simultaneously. We validate the proposed method on real-world clinical ultrasound videos of scanning the fetal head. The results demonstrate the feasibility of predicting ultrasound skills by the proposed framework, which quantifies optimised task performance as a skill indicator.
Neural Collapse-Inspired Multi-Label Federated Learning under Label-Distribution Skew
Sep 16, 2025Abstract:Federated Learning (FL) enables collaborative model training across distributed clients while preserving data privacy. However, the performance of deep learning often deteriorates in FL due to decentralized and heterogeneous data. This challenge is further amplified in multi-label scenarios, where data exhibit complex characteristics such as label co-occurrence, inter-label dependency, and discrepancies between local and global label relationships. While most existing FL research primarily focuses on single-label classification, many real-world applications, particularly in domains such as medical imaging, often involve multi-label settings. In this paper, we address this important yet underexplored scenario in FL, where clients hold multi-label data with skewed label distributions. Neural Collapse (NC) describes a geometric structure in the latent feature space where features of each class collapse to their class mean with vanishing intra-class variance, and the class means form a maximally separated configuration. Motivated by this theory, we propose a method to align feature distributions across clients and to learn high-quality, well-clustered representations. To make the NC-structure applicable to multi-label settings, where image-level features may contain multiple semantic concepts, we introduce a feature disentanglement module that extracts semantically specific features. The clustering of these disentangled class-wise features is guided by a predefined shared NC structure, which mitigates potential conflicts between client models due to diverse local data distributions. In addition, we design regularisation losses to encourage compact clustering in the latent feature space. Experiments conducted on four benchmark datasets across eight diverse settings demonstrate that our approach outperforms existing methods, validating its effectiveness in this challenging FL scenario.
Federated Continual 3D Segmentation With Single-round Communication
Mar 19, 2025



Abstract:Federated learning seeks to foster collaboration among distributed clients while preserving the privacy of their local data. Traditionally, federated learning methods assume a fixed setting in which client data and learning objectives remain constant. However, in real-world scenarios, new clients may join, and existing clients may expand the segmentation label set as task requirements evolve. In such a dynamic federated analysis setup, the conventional federated communication strategy of model aggregation per communication round is suboptimal. As new clients join, this strategy requires retraining, linearly increasing communication and computation overhead. It also imposes requirements for synchronized communication, which is difficult to achieve among distributed clients. In this paper, we propose a federated continual learning strategy that employs a one-time model aggregation at the server through multi-model distillation. This approach builds and updates the global model while eliminating the need for frequent server communication. When integrating new data streams or onboarding new clients, this approach efficiently reuses previous client models, avoiding the need to retrain the global model across the entire federation. By minimizing communication load and bypassing the need to put unchanged clients online, our approach relaxes synchronization requirements among clients, providing an efficient and scalable federated analysis framework suited for real-world applications. Using multi-class 3D abdominal CT segmentation as an application task, we demonstrate the effectiveness of the proposed approach.
Tell2Reg: Establishing spatial correspondence between images by the same language prompts
Feb 05, 2025Abstract:Spatial correspondence can be represented by pairs of segmented regions, such that the image registration networks aim to segment corresponding regions rather than predicting displacement fields or transformation parameters. In this work, we show that such a corresponding region pair can be predicted by the same language prompt on two different images using the pre-trained large multimodal models based on GroundingDINO and SAM. This enables a fully automated and training-free registration algorithm, potentially generalisable to a wide range of image registration tasks. In this paper, we present experimental results using one of the challenging tasks, registering inter-subject prostate MR images, which involves both highly variable intensity and morphology between patients. Tell2Reg is training-free, eliminating the need for costly and time-consuming data curation and labelling that was previously required for this registration task. This approach outperforms unsupervised learning-based registration methods tested, and has a performance comparable to weakly-supervised methods. Additional qualitative results are also presented to suggest that, for the first time, there is a potential correlation between language semantics and spatial correspondence, including the spatial invariance in language-prompted regions and the difference in language prompts between the obtained local and global correspondences. Code is available at https://github.com/yanwenCi/Tell2Reg.git.
AI-assisted prostate cancer detection and localisation on biparametric MR by classifying radiologist-positives
Oct 30, 2024



Abstract:Prostate cancer diagnosis through MR imaging have currently relied on radiologists' interpretation, whilst modern AI-based methods have been developed to detect clinically significant cancers independent of radiologists. In this study, we propose to develop deep learning models that improve the overall cancer diagnostic accuracy, by classifying radiologist-identified patients or lesions (i.e. radiologist-positives), as opposed to the existing models that are trained to discriminate over all patients. We develop a single voxel-level classification model, with a simple percentage threshold to determine positive cases, at levels of lesions, Barzell-zones and patients. Based on the presented experiments from two clinical data sets, consisting of histopathology-labelled MR images from more than 800 and 500 patients in the respective UCLA and UCL PROMIS studies, we show that the proposed strategy can improve the diagnostic accuracy, by augmenting the radiologist reading of the MR imaging. Among varying definition of clinical significance, the proposed strategy, for example, achieved a specificity of 44.1% (with AI assistance) from 36.3% (by radiologists alone), at a controlled sensitivity of 80.0% on the publicly available UCLA data set. This provides measurable clinical values in a range of applications such as reducing unnecessary biopsies, lowering cost in cancer screening and quantifying risk in therapies.
Nonrigid Reconstruction of Freehand Ultrasound without a Tracker
Jul 08, 2024Abstract:Reconstructing 2D freehand Ultrasound (US) frames into 3D space without using a tracker has recently seen advances with deep learning. Predicting good frame-to-frame rigid transformations is often accepted as the learning objective, especially when the ground-truth labels from spatial tracking devices are inherently rigid transformations. Motivated by a) the observed nonrigid deformation due to soft tissue motion during scanning, and b) the highly sensitive prediction of rigid transformation, this study investigates the methods and their benefits in predicting nonrigid transformations for reconstructing 3D US. We propose a novel co-optimisation algorithm for simultaneously estimating rigid transformations among US frames, supervised by ground-truth from a tracker, and a nonrigid deformation, optimised by a regularised registration network. We show that these two objectives can be either optimised using meta-learning or combined by weighting. A fast scattered data interpolation is also developed for enabling frequent reconstruction and registration of non-parallel US frames, during training. With a new data set containing over 357,000 frames in 720 scans, acquired from 60 subjects, the experiments demonstrate that, due to an expanded thus easier-to-optimise solution space, the generalisation is improved with the added deformation estimation, with respect to the rigid ground-truth. The global pixel reconstruction error (assessing accumulative prediction) is lowered from 18.48 to 16.51 mm, compared with baseline rigid-transformation-predicting methods. Using manually identified landmarks, the proposed co-optimisation also shows potentials in compensating nonrigid tissue motion at inference, which is not measurable by tracker-provided ground-truth. The code and data used in this paper are made publicly available at https://github.com/QiLi111/NR-Rec-FUS.
Semi-weakly-supervised neural network training for medical image registration
Feb 16, 2024



Abstract:For training registration networks, weak supervision from segmented corresponding regions-of-interest (ROIs) have been proven effective for (a) supplementing unsupervised methods, and (b) being used independently in registration tasks in which unsupervised losses are unavailable or ineffective. This correspondence-informing supervision entails cost in annotation that requires significant specialised effort. This paper describes a semi-weakly-supervised registration pipeline that improves the model performance, when only a small corresponding-ROI-labelled dataset is available, by exploiting unlabelled image pairs. We examine two types of augmentation methods by perturbation on network weights and image resampling, such that consistency-based unsupervised losses can be applied on unlabelled data. The novel WarpDDF and RegCut approaches are proposed to allow commutative perturbation between an image pair and the predicted spatial transformation (i.e. respective input and output of registration networks), distinct from existing perturbation methods for classification or segmentation. Experiments using 589 male pelvic MR images, labelled with eight anatomical ROIs, show the improvement in registration performance and the ablated contributions from the individual strategies. Furthermore, this study attempts to construct one of the first computational atlases for pelvic structures, enabled by registering inter-subject MRs, and quantifies the significant differences due to the proposed semi-weak supervision with a discussion on the potential clinical use of example atlas-derived statistics.
Boundary-RL: Reinforcement Learning for Weakly-Supervised Prostate Segmentation in TRUS Images
Aug 22, 2023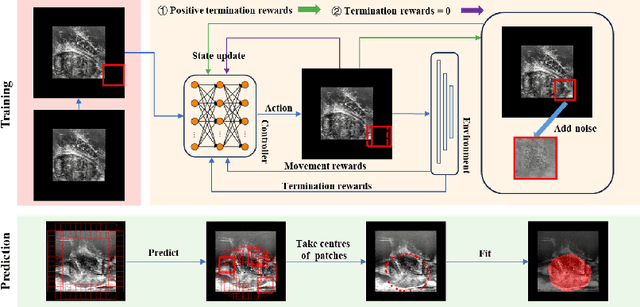
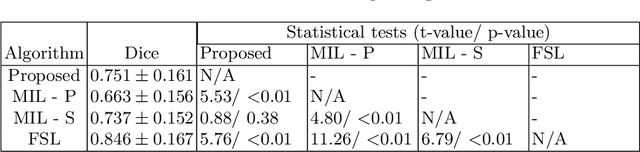
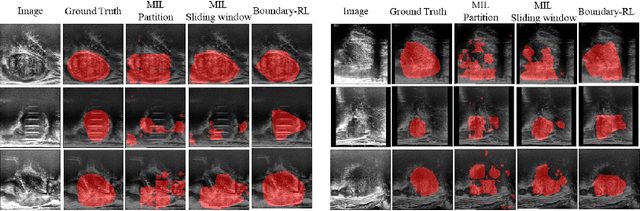
Abstract:We propose Boundary-RL, a novel weakly supervised segmentation method that utilises only patch-level labels for training. We envision the segmentation as a boundary detection problem, rather than a pixel-level classification as in previous works. This outlook on segmentation may allow for boundary delineation under challenging scenarios such as where noise artefacts may be present within the region-of-interest (ROI) boundaries, where traditional pixel-level classification-based weakly supervised methods may not be able to effectively segment the ROI. Particularly of interest, ultrasound images, where intensity values represent acoustic impedance differences between boundaries, may also benefit from the boundary delineation approach. Our method uses reinforcement learning to train a controller function to localise boundaries of ROIs using a reward derived from a pre-trained boundary-presence classifier. The classifier indicates when an object boundary is encountered within a patch, as the controller modifies the patch location in a sequential Markov decision process. The classifier itself is trained using only binary patch-level labels of object presence, which are the only labels used during training of the entire boundary delineation framework, and serves as a weak signal to inform the boundary delineation. The use of a controller function ensures that a sliding window over the entire image is not necessary. It also prevents possible false-positive or -negative cases by minimising number of patches passed to the boundary-presence classifier. We evaluate our proposed approach for a clinically relevant task of prostate gland segmentation on trans-rectal ultrasound images. We show improved performance compared to other tested weakly supervised methods, using the same labels e.g., multiple instance learning.
Combiner and HyperCombiner Networks: Rules to Combine Multimodality MR Images for Prostate Cancer Localisation
Jul 17, 2023



Abstract:One of the distinct characteristics in radiologists' reading of multiparametric prostate MR scans, using reporting systems such as PI-RADS v2.1, is to score individual types of MR modalities, T2-weighted, diffusion-weighted, and dynamic contrast-enhanced, and then combine these image-modality-specific scores using standardised decision rules to predict the likelihood of clinically significant cancer. This work aims to demonstrate that it is feasible for low-dimensional parametric models to model such decision rules in the proposed Combiner networks, without compromising the accuracy of predicting radiologic labels: First, it is shown that either a linear mixture model or a nonlinear stacking model is sufficient to model PI-RADS decision rules for localising prostate cancer. Second, parameters of these (generalised) linear models are proposed as hyperparameters, to weigh multiple networks that independently represent individual image modalities in the Combiner network training, as opposed to end-to-end modality ensemble. A HyperCombiner network is developed to train a single image segmentation network that can be conditioned on these hyperparameters during inference, for much improved efficiency. Experimental results based on data from 850 patients, for the application of automating radiologist labelling multi-parametric MR, compare the proposed combiner networks with other commonly-adopted end-to-end networks. Using the added advantages of obtaining and interpreting the modality combining rules, in terms of the linear weights or odds-ratios on individual image modalities, three clinical applications are presented for prostate cancer segmentation, including modality availability assessment, importance quantification and rule discovery.
Spatial Correspondence between Graph Neural Network-Segmented Images
Mar 17, 2023Abstract:Graph neural networks (GNNs) have been proposed for medical image segmentation, by predicting anatomical structures represented by graphs of vertices and edges. One such type of graph is predefined with fixed size and connectivity to represent a reference of anatomical regions of interest, thus known as templates. This work explores the potentials in these GNNs with common topology for establishing spatial correspondence, implicitly maintained during segmenting two or more images. With an example application of registering local vertebral sub-regions found in CT images, our experimental results showed that the GNN-based segmentation is capable of accurate and reliable localization of the same interventionally interesting structures between images, not limited to the segmentation classes. The reported average target registration errors of 2.2$\pm$1.3 mm and 2.7$\pm$1.4 mm, for aligning holdout test images with a reference and for aligning two test images, respectively, were by a considerable margin lower than those from the tested non-learning and learning-based registration algorithms. Further ablation studies assess the contributions towards the registration performance, from individual components in the originally segmentation-purposed network and its training algorithm. The results highlight that the proposed segmentation-in-lieu-of-registration approach shares methodological similarities with existing registration methods, such as the use of displacement smoothness constraint and point distance minimization albeit on non-grid graphs, which interestingly yielded benefits for both segmentation and registration. We, therefore, conclude that the template-based GNN segmentation can effectively establish spatial correspondence in our application, without any other dedicated registration algorithms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge