Verónica Vilaplana
Cell Nuclei Detection and Classification in Whole Slide Images with Transformers
Feb 10, 2025Abstract:Accurate and efficient cell nuclei detection and classification in histopathological Whole Slide Images (WSIs) are pivotal for digital pathology applications. Traditional cell segmentation approaches, while commonly used, are computationally expensive and require extensive post-processing, limiting their practicality for high-throughput clinical settings. In this paper, we propose a paradigm shift from segmentation to detection for extracting cell information from WSIs, introducing CellNuc-DETR as a more effective solution. We evaluate the accuracy performance of CellNuc-DETR on the PanNuke dataset and conduct cross-dataset evaluations on CoNSeP and MoNuSeg to assess robustness and generalization capabilities. Our results demonstrate state-of-the-art performance in both cell nuclei detection and classification tasks. Additionally, we assess the efficiency of CellNuc-DETR on large WSIs, showing that it not only outperforms current methods in accuracy but also significantly reduces inference times. Specifically, CellNuc-DETR is twice as fast as the fastest segmentation-based method, HoVer-NeXt, while achieving substantially higher accuracy. Moreover, it surpasses CellViT in accuracy and is approximately ten times more efficient in inference speed on WSIs. These results establish CellNuc-DETR as a superior approach for cell analysis in digital pathology, combining high accuracy with computational efficiency.
Layer-wise training for self-supervised learning on graphs
Sep 04, 2023Abstract:End-to-end training of graph neural networks (GNN) on large graphs presents several memory and computational challenges, and limits the application to shallow architectures as depth exponentially increases the memory and space complexities. In this manuscript, we propose Layer-wise Regularized Graph Infomax, an algorithm to train GNNs layer by layer in a self-supervised manner. We decouple the feature propagation and feature transformation carried out by GNNs to learn node representations in order to derive a loss function based on the prediction of future inputs. We evaluate the algorithm in inductive large graphs and show similar performance to other end to end methods and a substantially increased efficiency, which enables the training of more sophisticated models in one single device. We also show that our algorithm avoids the oversmoothing of the representations, another common challenge of deep GNNs.
RGI : Regularized Graph Infomax for self-supervised learning on graphs
Mar 15, 2023Abstract:Self-supervised learning is gaining considerable attention as a solution to avoid the requirement of extensive annotations in representation learning on graphs. We introduce \textit{Regularized Graph Infomax (RGI)}, a simple yet effective framework for node level self-supervised learning on graphs that trains a graph neural network encoder by maximizing the mutual information between node level local and global views, in contrast to previous works that employ graph level global views. The method promotes the predictability between views while regularizing the covariance matrices of the representations. Therefore, RGI is non-contrastive, does not depend on complex asymmetric architectures nor training tricks, is augmentation-free and does not rely on a two branch architecture. We run RGI on both transductive and inductive settings with popular graph benchmarks and show that it can achieve state-of-the-art performance regardless of its simplicity.
SurvLIMEpy: A Python package implementing SurvLIME
Feb 21, 2023Abstract:In this paper we present SurvLIMEpy, an open-source Python package that implements the SurvLIME algorithm. This method allows to compute local feature importance for machine learning algorithms designed for modelling Survival Analysis data. Our implementation takes advantage of the parallelisation paradigm as all computations are performed in a matrix-wise fashion which speeds up execution time. Additionally, SurvLIMEpy assists the user with visualization tools to better understand the result of the algorithm. The package supports a wide variety of survival models, from the Cox Proportional Hazards Model to deep learning models such as DeepHit or DeepSurv. Two types of experiments are presented in this paper. First, by means of simulated data, we study the ability of the algorithm to capture the importance of the features. Second, we use three open source survival datasets together with a set of survival algorithms in order to demonstrate how SurvLIMEpy behaves when applied to different models.
Standardized Assessment of Automatic Segmentation of White Matter Hyperintensities and Results of the WMH Segmentation Challenge
Apr 01, 2019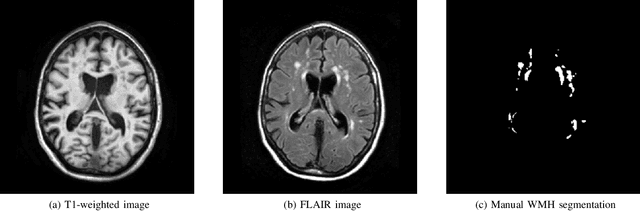
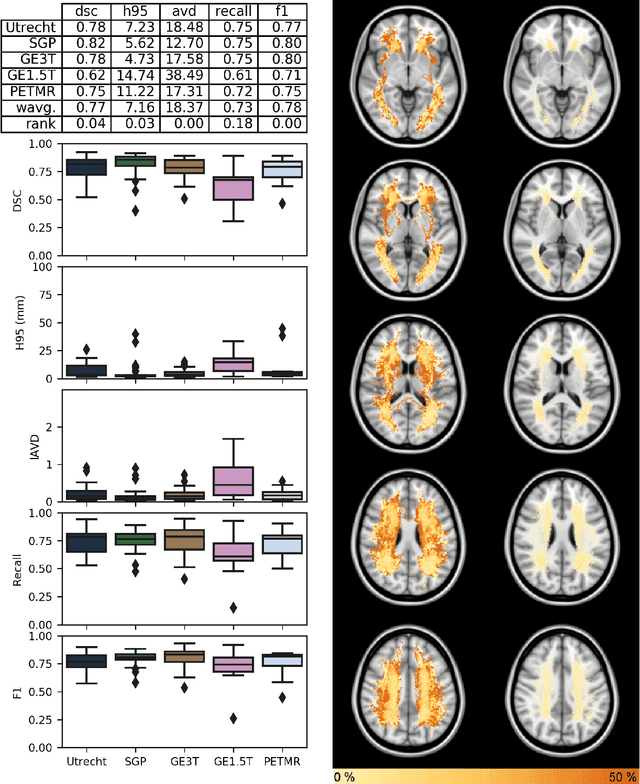
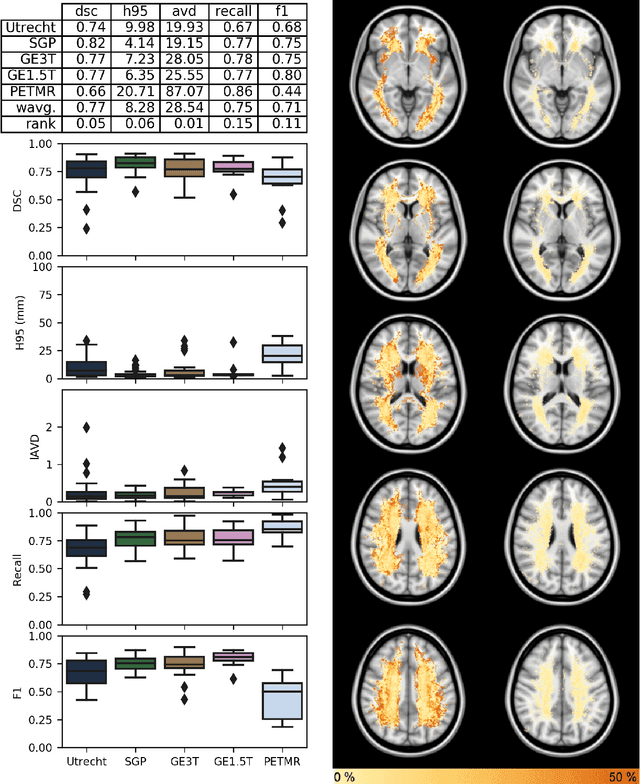
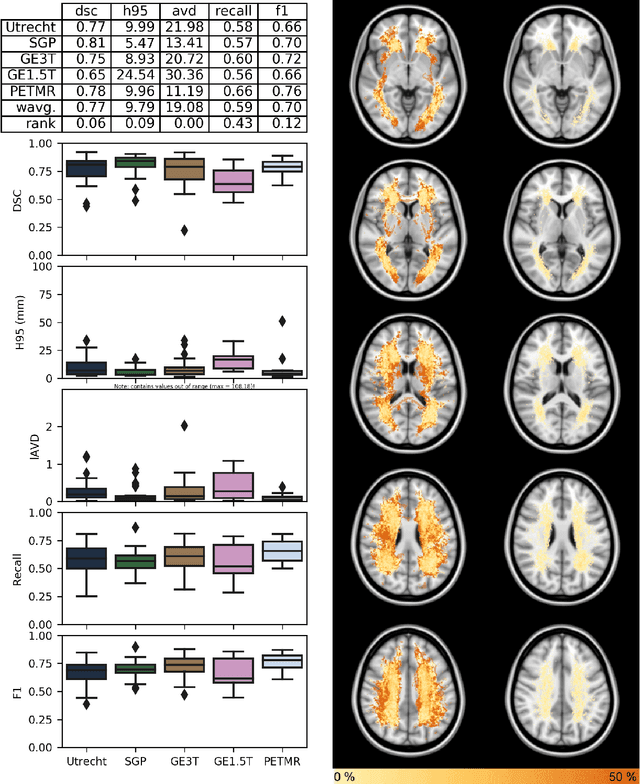
Abstract:Quantification of cerebral white matter hyperintensities (WMH) of presumed vascular origin is of key importance in many neurological research studies. Currently, measurements are often still obtained from manual segmentations on brain MR images, which is a laborious procedure. Automatic WMH segmentation methods exist, but a standardized comparison of the performance of such methods is lacking. We organized a scientific challenge, in which developers could evaluate their method on a standardized multi-center/-scanner image dataset, giving an objective comparison: the WMH Segmentation Challenge (https://wmh.isi.uu.nl/). Sixty T1+FLAIR images from three MR scanners were released with manual WMH segmentations for training. A test set of 110 images from five MR scanners was used for evaluation. Segmentation methods had to be containerized and submitted to the challenge organizers. Five evaluation metrics were used to rank the methods: (1) Dice similarity coefficient, (2) modified Hausdorff distance (95th percentile), (3) absolute log-transformed volume difference, (4) sensitivity for detecting individual lesions, and (5) F1-score for individual lesions. Additionally, methods were ranked on their inter-scanner robustness. Twenty participants submitted their method for evaluation. This paper provides a detailed analysis of the results. In brief, there is a cluster of four methods that rank significantly better than the other methods, with one clear winner. The inter-scanner robustness ranking shows that not all methods generalize to unseen scanners. The challenge remains open for future submissions and provides a public platform for method evaluation.
Cascaded V-Net using ROI masks for brain tumor segmentation
Dec 30, 2018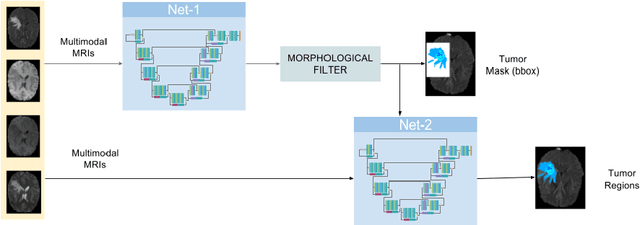
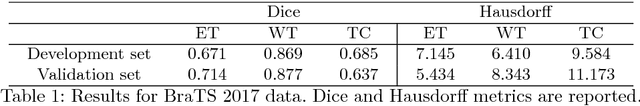
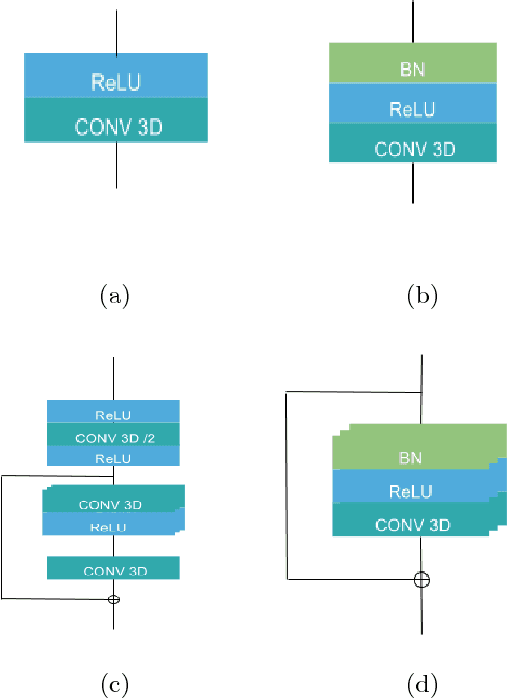
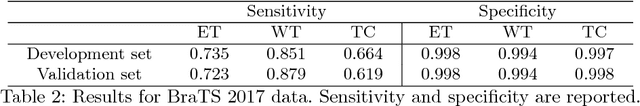
Abstract:In this work we approach the brain tumor segmentation problem with a cascade of two CNNs inspired in the V-Net architecture \cite{VNet}, reformulating residual connections and making use of ROI masks to constrain the networks to train only on relevant voxels. This architecture allows dense training on problems with highly skewed class distributions, such as brain tumor segmentation, by focusing training only on the vecinity of the tumor area. We report results on BraTS2017 Training and Validation sets.
* Third International Workshop, BrainLes 2017, Held in Conjunction with MICCAI 2017, Quebec City, QC, Canada, September 14, 2017, Revised Selected Papers
Leishmaniasis Parasite Segmentation and Classification using Deep Learning
Dec 30, 2018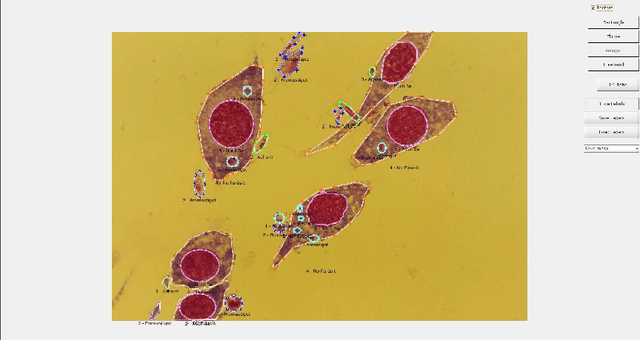
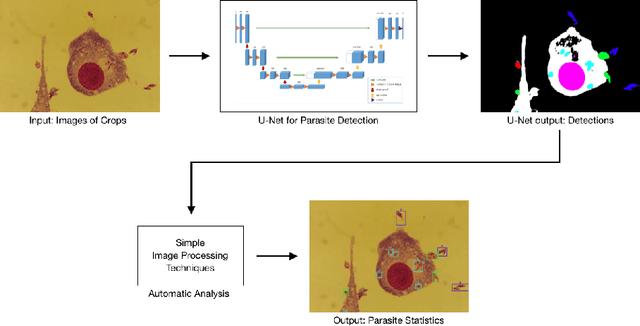
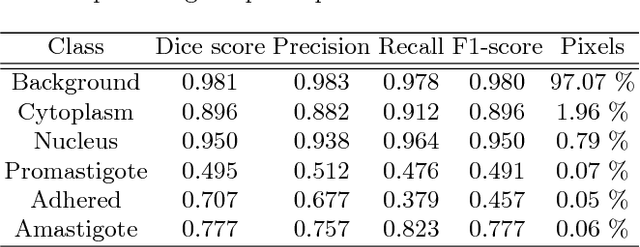

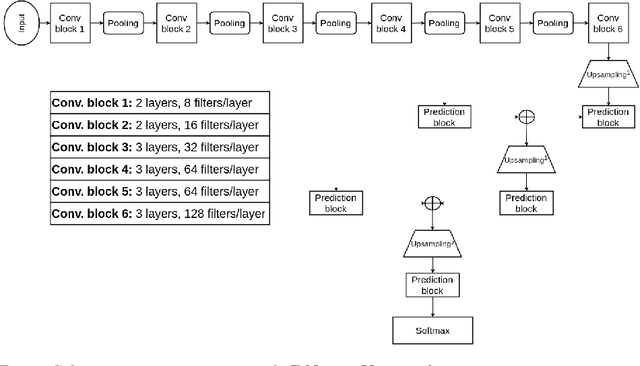
Abstract:Leishmaniasis is considered a neglected disease that causes thousands of deaths annually in some tropical and subtropical countries. There are various techniques to diagnose leishmaniasis of which manual microscopy is considered to be the gold standard. There is a need for the development of automatic techniques that are able to detect parasites in a robust and unsupervised manner. In this paper we present a procedure for automatizing the detection process based on a deep learning approach. We train a U-net model that successfully segments leismania parasites and classifies them into promastigotes, amastigotes and adhered parasites.
* 10th International Conference, AMDO 2018, Palma de Mallorca, Spain, July 12-13, 2018, Proceedings
3D Convolutional Neural Networks for Brain Tumor Segmentation: A Comparison of Multi-resolution Architectures
May 23, 2017


Abstract:This paper analyzes the use of 3D Convolutional Neural Networks for brain tumor segmentation in MR images. We address the problem using three different architectures that combine fine and coarse features to obtain the final segmentation. We compare three different networks that use multi-resolution features in terms of both design and performance and we show that they improve their single-resolution counterparts.
Saliency maps on image hierarchies
Aug 19, 2015



Abstract:In this paper we propose two saliency models for salient object segmentation based on a hierarchical image segmentation, a tree-like structure that represents regions at different scales from the details to the whole image (e.g. gPb-UCM, BPT). The first model is based on a hierarchy of image partitions. The saliency at each level is computed on a region basis, taking into account the contrast between regions. The maps obtained for the different partitions are then integrated into a final saliency map. The second model directly works on the structure created by the segmentation algorithm, computing saliency at each node and integrating these cues in a straightforward manner into a single saliency map. We show that the proposed models produce high quality saliency maps. Objective evaluation demonstrates that the two methods achieve state-of-the-art performance in several benchmark datasets.
Improving Spatial Codification in Semantic Segmentation
May 27, 2015
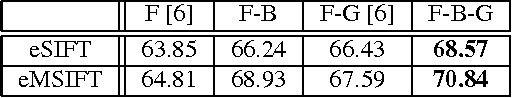
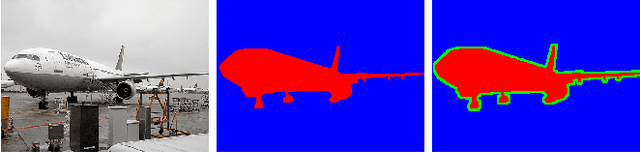
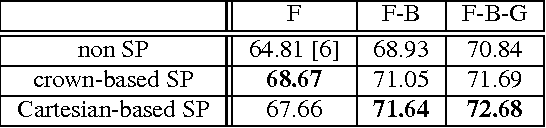
Abstract:This paper explores novel approaches for improving the spatial codification for the pooling of local descriptors to solve the semantic segmentation problem. We propose to partition the image into three regions for each object to be described: Figure, Border and Ground. This partition aims at minimizing the influence of the image context on the object description and vice versa by introducing an intermediate zone around the object contour. Furthermore, we also propose a richer visual descriptor of the object by applying a Spatial Pyramid over the Figure region. Two novel Spatial Pyramid configurations are explored: Cartesian-based and crown-based Spatial Pyramids. We test these approaches with state-of-the-art techniques and show that they improve the Figure-Ground based pooling in the Pascal VOC 2011 and 2012 semantic segmentation challenges.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge