Mariana Bento
Department of Biomedical Engineering, University of Calgary, Calgary, Canada, Hotchkiss Brain Institute, University of Calgary, Calgary, Canada, Department of Electrical and Software Engineering, University of Calgary, Calgary, Canada
Trustworthy and Responsible AI for Human-Centric Autonomous Decision-Making Systems
Sep 02, 2024Abstract:Artificial Intelligence (AI) has paved the way for revolutionary decision-making processes, which if harnessed appropriately, can contribute to advancements in various sectors, from healthcare to economics. However, its black box nature presents significant ethical challenges related to bias and transparency. AI applications are hugely impacted by biases, presenting inconsistent and unreliable findings, leading to significant costs and consequences, highlighting and perpetuating inequalities and unequal access to resources. Hence, developing safe, reliable, ethical, and Trustworthy AI systems is essential. Our team of researchers working with Trustworthy and Responsible AI, part of the Transdisciplinary Scholarship Initiative within the University of Calgary, conducts research on Trustworthy and Responsible AI, including fairness, bias mitigation, reproducibility, generalization, interpretability, and authenticity. In this paper, we review and discuss the intricacies of AI biases, definitions, methods of detection and mitigation, and metrics for evaluating bias. We also discuss open challenges with regard to the trustworthiness and widespread application of AI across diverse domains of human-centric decision making, as well as guidelines to foster Responsible and Trustworthy AI models.
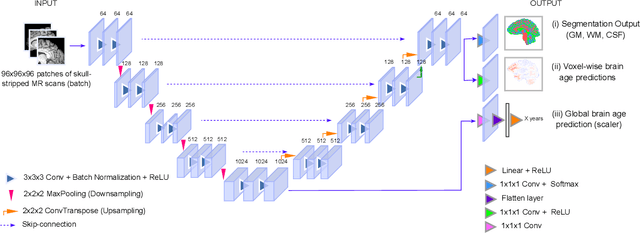
A voxel-level approach to brain age prediction: A method to assess regional brain aging
Oct 17, 2023



Abstract:Brain aging is a regional phenomenon, a facet that remains relatively under-explored within the realm of brain age prediction research using machine learning methods. Voxel-level predictions can provide localized brain age estimates that can provide granular insights into the regional aging processes. This is essential to understand the differences in aging trajectories in healthy versus diseased subjects. In this work, a deep learning-based multitask model is proposed for voxel-level brain age prediction from T1-weighted magnetic resonance images. The proposed model outperforms the models existing in the literature and yields valuable clinical insights when applied to both healthy and diseased populations. Regional analysis is performed on the voxel-level brain age predictions to understand aging trajectories of known anatomical regions in the brain and show that there exist disparities in regional aging trajectories of healthy subjects compared to ones with underlying neurological disorders such as Dementia and more specifically, Alzheimer's disease. Our code is available at https://github.com/nehagianchandani/Voxel-level-brain-age-prediction.
Studying the Effects of Sex-related Differences on Brain Age Prediction using brain MR Imaging
Oct 17, 2023Abstract:While utilizing machine learning models, one of the most crucial aspects is how bias and fairness affect model outcomes for diverse demographics. This becomes especially relevant in the context of machine learning for medical imaging applications as these models are increasingly being used for diagnosis and treatment planning. In this paper, we study biases related to sex when developing a machine learning model based on brain magnetic resonance images (MRI). We investigate the effects of sex by performing brain age prediction considering different experimental designs: model trained using only female subjects, only male subjects and a balanced dataset. We also perform evaluation on multiple MRI datasets (Calgary-Campinas(CC359) and CamCAN) to assess the generalization capability of the proposed models. We found disparities in the performance of brain age prediction models when trained on distinct sex subgroups and datasets, in both final predictions and decision making (assessed using interpretability models). Our results demonstrated variations in model generalizability across sex-specific subgroups, suggesting potential biases in models trained on unbalanced datasets. This underlines the critical role of careful experimental design in generating fair and reliable outcomes.
Reframing the Brain Age Prediction Problem to a More Interpretable and Quantitative Approach
Aug 23, 2023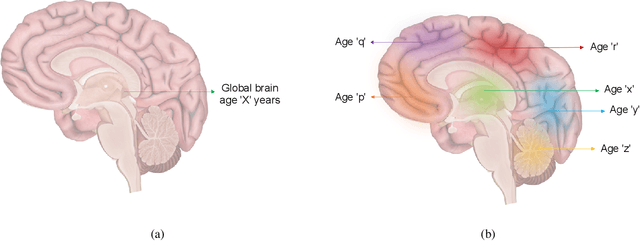

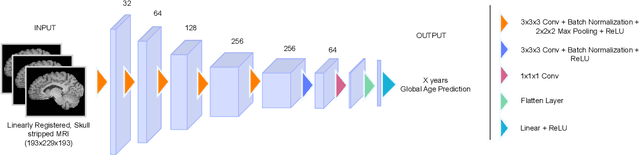
Abstract:Deep learning models have achieved state-of-the-art results in estimating brain age, which is an important brain health biomarker, from magnetic resonance (MR) images. However, most of these models only provide a global age prediction, and rely on techniques, such as saliency maps to interpret their results. These saliency maps highlight regions in the input image that were significant for the model's predictions, but they are hard to be interpreted, and saliency map values are not directly comparable across different samples. In this work, we reframe the age prediction problem from MR images to an image-to-image regression problem where we estimate the brain age for each brain voxel in MR images. We compare voxel-wise age prediction models against global age prediction models and their corresponding saliency maps. The results indicate that voxel-wise age prediction models are more interpretable, since they provide spatial information about the brain aging process, and they benefit from being quantitative.
Dual-domain Cascade of U-nets for Multi-channel Magnetic Resonance Image Reconstruction
Nov 04, 2019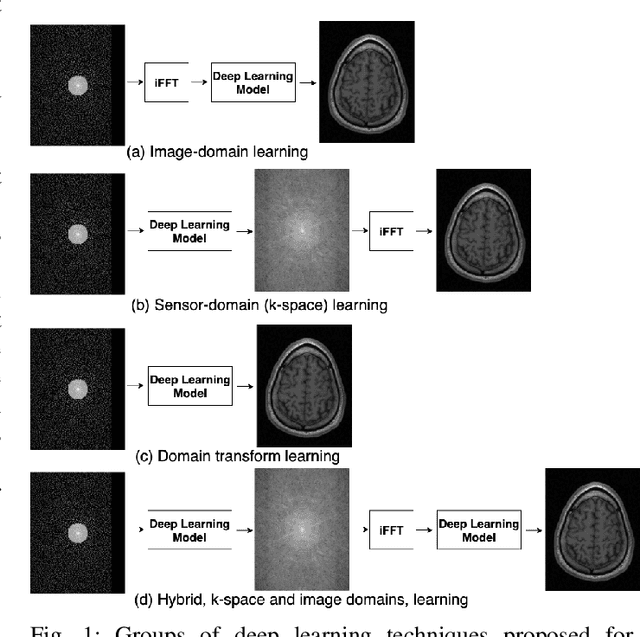
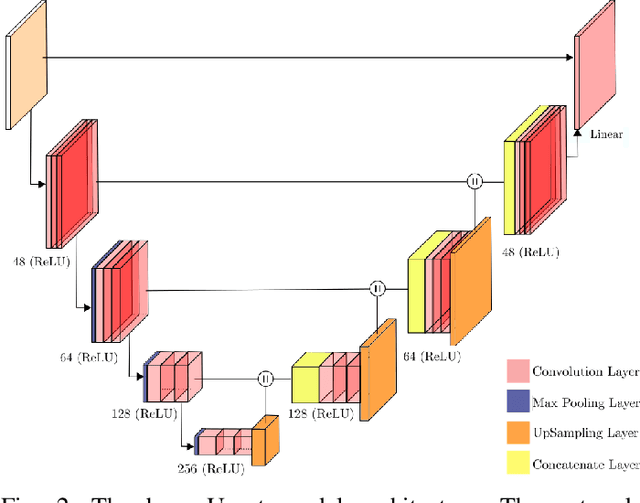
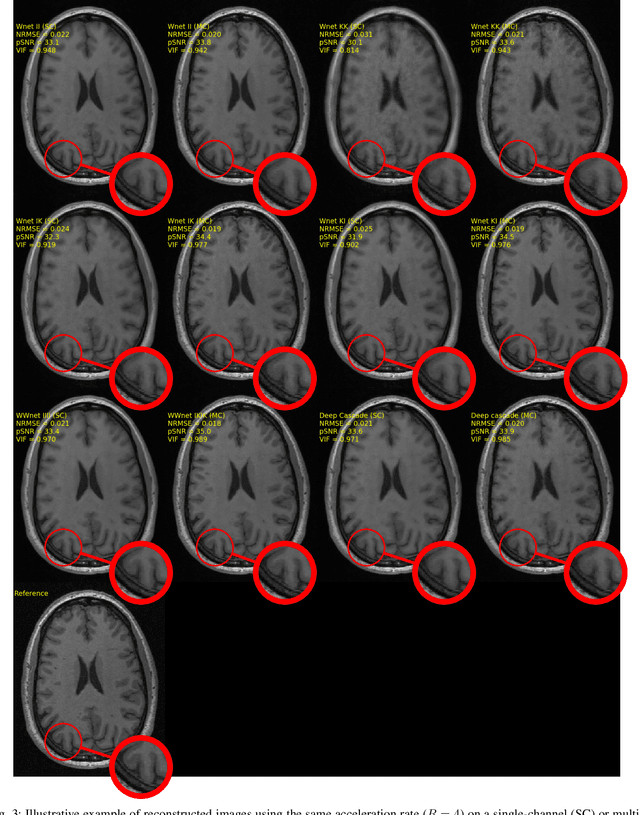
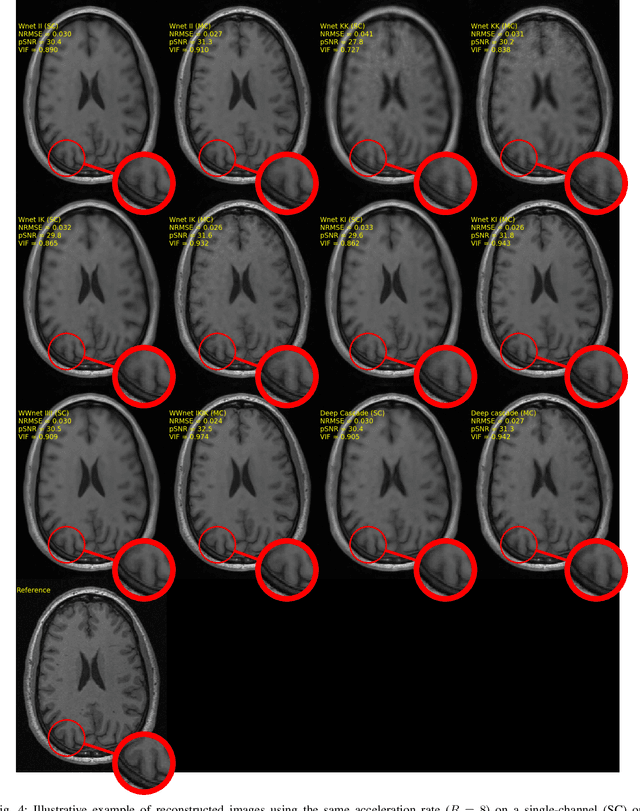
Abstract:The U-net is a deep-learning network model that has been used to solve a number of inverse problems. In this work, the concatenation of two-element U-nets, termed the W-net, operating in k-space (K) and image (I) domains, were evaluated for multi-channel magnetic resonance (MR) image reconstruction. The two element network combinations were evaluated for the four possible image-k-space domain configurations: a) W-net II, b) W-net KK, c) W-net IK, and d) W-net KI were evaluated. Selected promising four element networks (WW-nets) were also examined. Two configurations of each network were compared: 1) Each coil channel processed independently, and 2) all channels processed simultaneously. One hundred and eleven volumetric, T1-weighted, 12-channel coil k-space datasets were used in the experiments. Normalized root mean squared error, peak signal to noise ratio, visual information fidelity and visual inspection were used to assess the reconstructed images against the fully sampled reference images. Our results indicated that networks that operate solely in the image domain are better suited when processing individual channels of multi-channel data independently. Dual domain methods are more advantageous when simultaneously reconstructing all channels of multi-channel data. Also, the appropriate cascade of U-nets compared favorably (p < 0.01) to the previously published, state-of-the-art Deep Cascade model in in three out of four experiments.
Standardized Assessment of Automatic Segmentation of White Matter Hyperintensities and Results of the WMH Segmentation Challenge
Apr 01, 2019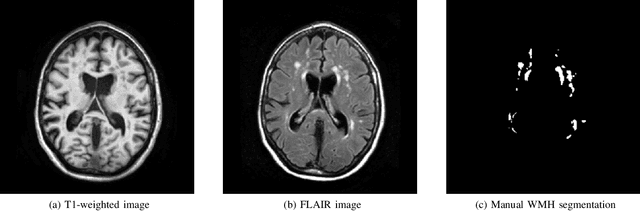
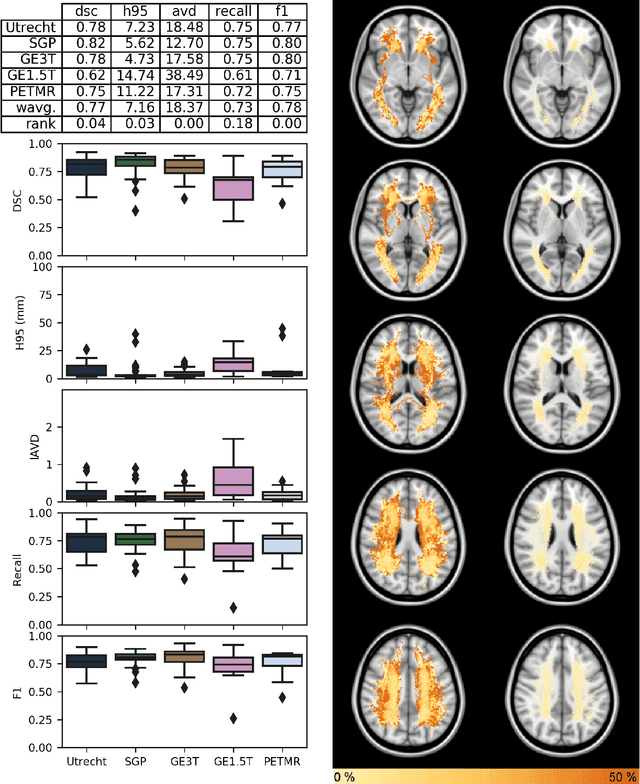
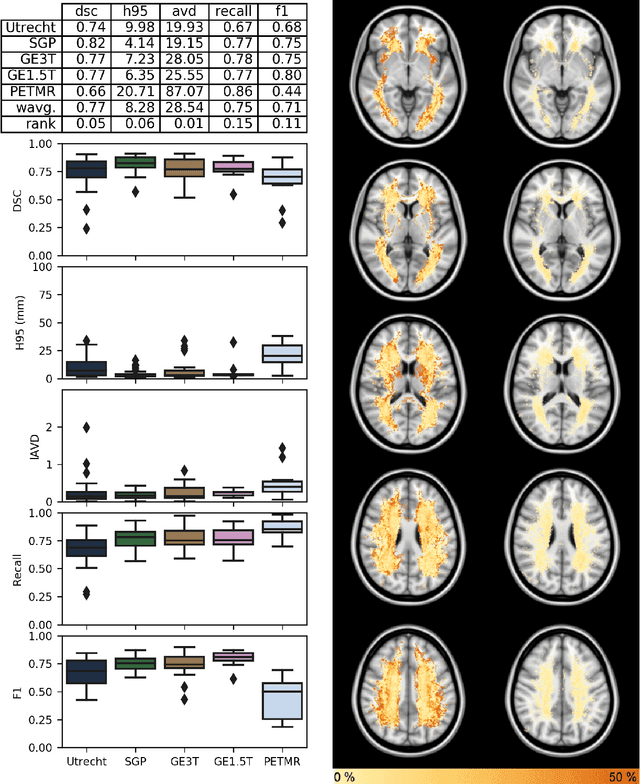
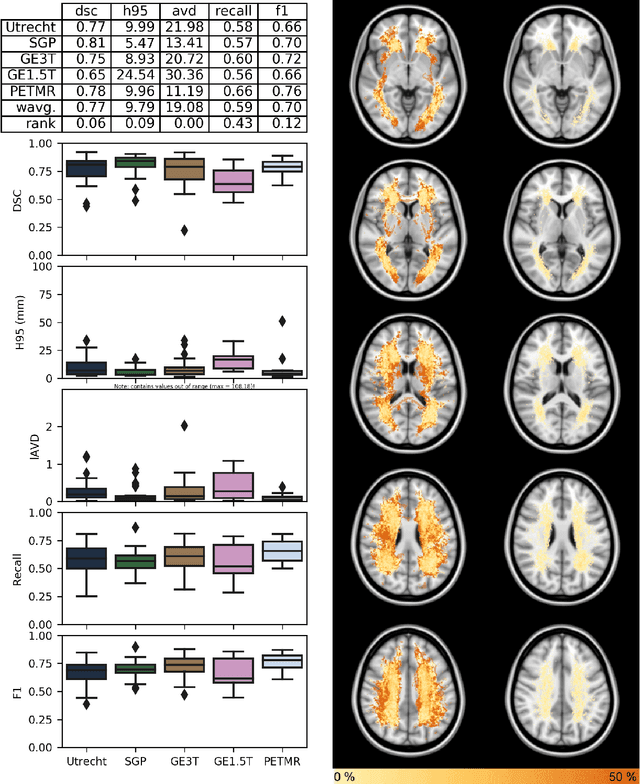
Abstract:Quantification of cerebral white matter hyperintensities (WMH) of presumed vascular origin is of key importance in many neurological research studies. Currently, measurements are often still obtained from manual segmentations on brain MR images, which is a laborious procedure. Automatic WMH segmentation methods exist, but a standardized comparison of the performance of such methods is lacking. We organized a scientific challenge, in which developers could evaluate their method on a standardized multi-center/-scanner image dataset, giving an objective comparison: the WMH Segmentation Challenge (https://wmh.isi.uu.nl/). Sixty T1+FLAIR images from three MR scanners were released with manual WMH segmentations for training. A test set of 110 images from five MR scanners was used for evaluation. Segmentation methods had to be containerized and submitted to the challenge organizers. Five evaluation metrics were used to rank the methods: (1) Dice similarity coefficient, (2) modified Hausdorff distance (95th percentile), (3) absolute log-transformed volume difference, (4) sensitivity for detecting individual lesions, and (5) F1-score for individual lesions. Additionally, methods were ranked on their inter-scanner robustness. Twenty participants submitted their method for evaluation. This paper provides a detailed analysis of the results. In brief, there is a cluster of four methods that rank significantly better than the other methods, with one clear winner. The inter-scanner robustness ranking shows that not all methods generalize to unseen scanners. The challenge remains open for future submissions and provides a public platform for method evaluation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge